Chromatin Bridge on:
[Wikipedia]
[Google]
[Amazon]
 Chromatin is a complex of
Chromatin is a complex of

 Chromatin undergoes various structural changes during a
Chromatin undergoes various structural changes during a
 The basic repeat element of chromatin is the nucleosome, interconnected by sections of linker DNA, a far shorter arrangement than pure DNA in solution.
In addition to core histones, a linker
The basic repeat element of chromatin is the nucleosome, interconnected by sections of linker DNA, a far shorter arrangement than pure DNA in solution.
In addition to core histones, a linker
 With addition of H1, during
With addition of H1, during  The existing models commonly accept that the nucleosomes lie perpendicular to the axis of the fibre, with linker histones arranged internally.
A stable 30 nm fibre relies on the regular positioning of nucleosomes along DNA. Linker DNA is relatively resistant to bending and rotation. This makes the length of linker DNA critical to the stability of the fibre, requiring nucleosomes to be separated by lengths that permit rotation and folding into the required orientation without excessive stress to the DNA.
In this view, different lengths of the linker DNA should produce different folding topologies of the chromatin fiber. Recent theoretical work, based on electron-microscopy images
of reconstituted fibers supports this view.
The existing models commonly accept that the nucleosomes lie perpendicular to the axis of the fibre, with linker histones arranged internally.
A stable 30 nm fibre relies on the regular positioning of nucleosomes along DNA. Linker DNA is relatively resistant to bending and rotation. This makes the length of linker DNA critical to the stability of the fibre, requiring nucleosomes to be separated by lengths that permit rotation and folding into the required orientation without excessive stress to the DNA.
In this view, different lengths of the linker DNA should produce different folding topologies of the chromatin fiber. Recent theoretical work, based on electron-microscopy images
of reconstituted fibers supports this view.

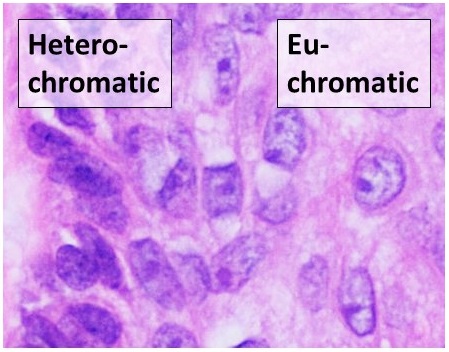
 # Interphase: The structure of chromatin during
# Interphase: The structure of chromatin during
 #
#
Chromosomes and Chromatin.
* * Cremer, T. 1985. Von der Zellenlehre zur Chromosomentheorie: Naturwissenschaftliche Erkenntnis und Theorienwechsel in der frühen Zell- und Vererbungsforschung, Veröffentlichungen aus der Forschungsstelle für Theoretische Pathologie der Heidelberger Akademie der Wissenschaften. Springer-Vlg., Berlin, Heidelberg. * Elgin, S. C. R. (ed.). 1995. Chromatin Structure and Gene Expression, vol. 9. IRL Press, Oxford, New York, Tokyo. * * * * * * Pollard, T., and W. Earnshaw. 2002. Cell Biology. Saunders. * Saumweber, H. 1987. Arrangement of Chromosomes in Interphase Cell Nuclei, p. 223-234. In W. Hennig (ed.), Structure and Function of Eucaryotic Chromosomes, vol. 14. Springer-Verlag, Berlin, Heidelberg. * * Van Holde KE. 1989. Chromatin. New York:
Chromatin, Histones & Cathepsin
PMAP The Proteolysis Map-animation
''Nature'' journal: recent chromatin publications and news
Protocol for ''in vitro'' Chromatin Assembly
ENCODE threads Explorer
Chromatin patterns at transcription factor binding sites.
 Chromatin is a complex of
Chromatin is a complex of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
and protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
found in eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division
Cell division is the process by which a parent cell (biology), cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukar ...
, preventing DNA damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified ...
, and regulating gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
and DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
. During mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
and meiosis
Meiosis () is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one c ...
, chromatin facilitates proper segregation of the chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
s in anaphase
Anaphase () is the stage of mitosis after the process of metaphase, when replicated chromosomes are split and the newly-copied chromosomes (daughter chromatids) are moved to opposite poles of the cell. Chromosomes also reach their overall maxim ...
; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.
The primary protein components of chromatin are histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
s. An octamer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomer, monomers.Quote: ''Oligomer molecule: A molecule of intermediate ...
of two sets of four histone cores (Histone H2A
Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells.
The other histone proteins are: Histone H1, H1, Histone H2B, H2B, Histone H3, H3 and Histone H4, H4.
Background
Histones are pro ...
, Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes.
Struc ...
, Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'b ...
, and Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleo ...
) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30 nm fiber. ''Current opinion in cell biology, 58,'' 95–104. https://doi.org/10.1016/j.ceb.2019.02.003 In general, there are three levels of chromatin organization:
# DNA wraps around histone proteins, forming nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
s and the so-called beads on a string
A bead is a small, decorative object that is formed in a variety of shapes and sizes of a material such as stone, bone, shell, glass, plastic, wood, or pearl and with a small hole for threading or stringing. Beads range in size from under 1 ...
structure (euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, which ...
).
# Multiple histones wrap into a 30-nanometer
330px, Different lengths as in respect to the Molecule">molecular scale.
The nanometre (international spelling as used by the International Bureau of Weights and Measures; SI symbol: nm), or nanometer (American spelling
Despite the va ...
fiber consisting of nucleosome arrays in their most compact form (heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
).
# Higher-level DNA supercoiling of the 30 nm fiber produces the metaphase
Metaphase ( and ) is a stage of mitosis in the eukaryotic cell cycle in which chromosomes are at their second-most condensed and coiled stage (they are at their most condensed in anaphase). These chromosomes, carrying genetic information, alig ...
chromosome (during mitosis and meiosis).
Many organisms, however, do not follow this organization scheme. For example, spermatozoa
A spermatozoon (; also spelled spermatozoön; : spermatozoa; ) is a motile sperm cell (biology), cell produced by male animals relying on internal fertilization. A spermatozoon is a moving form of the ploidy, haploid cell (biology), cell that is ...
and avian red blood cell
Red blood cells (RBCs), referred to as erythrocytes (, with -''cyte'' translated as 'cell' in modern usage) in academia and medical publishing, also known as red cells, erythroid cells, and rarely haematids, are the most common type of blood cel ...
s have more tightly packed chromatin than most eukaryotic cells, and trypanosomatid
Trypanosomatida is a group of kinetoplastid unicellular organisms distinguished by having only a single flagellum. The name is derived from the Greek ''trypano'' (borer) and ''soma'' (body) because of the corkscrew-like motion of some trypanosoma ...
protozoa
Protozoa (: protozoan or protozoon; alternative plural: protozoans) are a polyphyletic group of single-celled eukaryotes, either free-living or parasitic, that feed on organic matter such as other microorganisms or organic debris. Historically ...
do not condense
Condensation is the change of the state of matter from the gas phase into the liquid phase, and is the reverse of vaporization. The word most often refers to the water cycle. It can also be defined as the change in the state of water vapor ...
their chromatin into visible chromosomes at all. Prokaryotic
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
cells have entirely different structures for organizing their DNA (the prokaryotic chromosome equivalent is called a genophore and is localized within the nucleoid region).
The overall structure of the chromatin network further depends on the stage of the cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
. During interphase
Interphase is the active portion of the cell cycle that includes the G1, S, and G2 phases, where the cell grows, replicates its DNA, and prepares for mitosis, respectively. Interphase was formerly called the "resting phase," but the cell i ...
, the chromatin is structurally loose to allow access to RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
and DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
s that transcribe and replicate the DNA. The local structure of chromatin during interphase depends on the specific gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
s present in the DNA. Regions of DNA containing genes which are actively transcribed ("turned on") are less tightly compacted and closely associated with RNA polymerases in a structure known as euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, which ...
, while regions containing inactive genes ("turned off") are generally more condensed and associated with structural proteins in heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
. Epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
modification of the structural proteins in chromatin via methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
and acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
also alters local chromatin structure and therefore gene expression. There is limited understanding of chromatin structure and it is active area of research in molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
.
Dynamic chromatin structure and hierarchy
cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
. Histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
proteins are the basic packers and arrangers of chromatin and can be modified by various post-translational modifications to alter chromatin packing (histone modification
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. ...
). Most modifications occur on histone tails. The positively charged histone cores only partially counteract the negative charge of the DNA phosphate backbone resulting in a negative net charge of the overall structure. An imbalance of charge within the polymer causes electrostatic
Electrostatics is a branch of physics that studies slow-moving or stationary electric charges.
Since classical times, it has been known that some materials, such as amber, attract lightweight particles after rubbing. The Greek word (), mean ...
repulsion between neighboring chromatin regions that promote interactions with positively charged proteins, molecules, and cations. As these modifications occur, the electrostatic environment surrounding the chromatin will flux and the level of chromatin compaction will alter. The consequences in terms of chromatin accessibility and compaction depend both on the modified amino acid and the type of modification. For example, histone acetylation results in loosening and increased accessibility of chromatin for replication and transcription. Lysine trimethylation can either lead to increased transcriptional activity ( trimethylation of histone H3 lysine 4) or transcriptional repression and chromatin compaction ( trimethylation of histone H3, lysine 9 or lysine 27). Several studies suggested that different modifications could occur simultaneously. For example, it was proposed that a bivalent structure (with trimethylation of both lysine 4 and 27 on histone H3) is involved in early mammalian development. Another study tested the role of acetylation of histone 4 on lysine 16 on chromatin structure and found that homogeneous
Homogeneity and heterogeneity are concepts relating to the uniformity of a substance, process or image. A homogeneous feature is uniform in composition or character (i.e., color, shape, size, weight, height, distribution, texture, language, i ...
acetylation inhibited 30 nm chromatin formation and blocked adenosine triphosphate
Adenosine triphosphate (ATP) is a nucleoside triphosphate that provides energy to drive and support many processes in living cell (biology), cells, such as muscle contraction, nerve impulse propagation, and chemical synthesis. Found in all known ...
remodeling. This singular modification changed the dynamics of the chromatin which shows that acetylation of H4 at K16 is vital for proper intra- and inter- functionality of chromatin structure.
Polycomb-group proteins
Polycomb-group proteins (PcG proteins) are a family of protein complexes first discovered in fruit flies that can remodel chromatin such that epigenetic silencing of genes takes place. Polycomb-group proteins are well known for silencing Hox genes ...
play a role in regulating genes through modulation of chromatin structure.
For additional information, see Chromatin variant, Histone modifications in chromatin regulation and RNA polymerase control by chromatin structure.
Structure of DNA
In nature, DNA can form three structures, A-, B-, and Z-DNA. A- and B-DNA are very similar, forming right-handed helices, whereas Z-DNA is a left-handed helix with a zig-zag phosphate backbone. Z-DNA is thought to play a specific role in chromatin structure and transcription because of the properties of the junction between B- and Z-DNA. At the junction of B- and Z-DNA, one pair of bases is flipped out from normal bonding. These play a dual role of a site of recognition by many proteins and as a sink for torsional stress fromRNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
or nucleosome binding. DNA bases are stored as a code structure with four chemical bases such as ''"Adenine (A), Guanine (G), Cytosine (C), and Thymine (T)"''. The order and sequences of these chemical structures of DNA are reflected as information available for the creation and control of human organisms. ''"A with T and C with G"'' pairing up to build the DNA base pair. ''Sugar and phosphate'' molecules are also paired with these bases, making DNA nucleotides arrange 2 long spiral strands unitedly called ''"double helix"''. In eukaryotes, DNA consists of a cell nucleus and the DNA is providing strength and direction to the mechanism of heredity. Moreover, between the nitrogenous bonds of the 2 DNA, homogenous bonds are forming.
Nucleosomes and beads-on-a-string
 The basic repeat element of chromatin is the nucleosome, interconnected by sections of linker DNA, a far shorter arrangement than pure DNA in solution.
In addition to core histones, a linker
The basic repeat element of chromatin is the nucleosome, interconnected by sections of linker DNA, a far shorter arrangement than pure DNA in solution.
In addition to core histones, a linker histone H1
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species.
Structure
Metazoan H1 prote ...
exists that contacts the exit/entry of the DNA strand on the nucleosome. The nucleosome core particle, together with histone H1, is known as a chromatosome
In molecular biology, a chromatosome is a result of histone H1 binding to a nucleosome, which contains a histone octamer and DNA. The chromatosome contains 166 base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids ...
. Nucleosomes, with about 20 to 60 base pairs of linker DNA, can form, under non-physiological conditions, an approximately 11 nm beads on a string
A bead is a small, decorative object that is formed in a variety of shapes and sizes of a material such as stone, bone, shell, glass, plastic, wood, or pearl and with a small hole for threading or stringing. Beads range in size from under 1 ...
fibre.
The nucleosomes bind DNA non-specifically, as required by their function in general DNA packaging. There are, however, large DNA sequence preferences that govern nucleosome positioning. This is due primarily to the varying physical properties of different DNA sequences: For instance, adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
(A), and thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
(T) is more favorably compressed into the inner minor grooves. This means nucleosomes can bind preferentially at one position approximately every 10 base pairs (the helical repeat of DNA)- where the DNA is rotated to maximise the number of A and T bases that will lie in the inner minor groove. (See nucleic acid structure.)
30-nm chromatin fiber in mitosis
 With addition of H1, during
With addition of H1, during mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
the beads-on-a-string structure can coil into a 30 nm-diameter helical structure known as the 30 nm fibre or filament. The precise structure of the chromatin fiber in the cell is not known in detail.
This level of chromatin structure is thought to be the form of heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
, which contains mostly transcriptionally silent genes. Electron microscopy studies have demonstrated that the 30 nm fiber is highly dynamic such that it unfolds into a 10 nm fiber beads-on-a-string structure when transversed by an RNA polymerase engaged in transcription.
 The existing models commonly accept that the nucleosomes lie perpendicular to the axis of the fibre, with linker histones arranged internally.
A stable 30 nm fibre relies on the regular positioning of nucleosomes along DNA. Linker DNA is relatively resistant to bending and rotation. This makes the length of linker DNA critical to the stability of the fibre, requiring nucleosomes to be separated by lengths that permit rotation and folding into the required orientation without excessive stress to the DNA.
In this view, different lengths of the linker DNA should produce different folding topologies of the chromatin fiber. Recent theoretical work, based on electron-microscopy images
of reconstituted fibers supports this view.
The existing models commonly accept that the nucleosomes lie perpendicular to the axis of the fibre, with linker histones arranged internally.
A stable 30 nm fibre relies on the regular positioning of nucleosomes along DNA. Linker DNA is relatively resistant to bending and rotation. This makes the length of linker DNA critical to the stability of the fibre, requiring nucleosomes to be separated by lengths that permit rotation and folding into the required orientation without excessive stress to the DNA.
In this view, different lengths of the linker DNA should produce different folding topologies of the chromatin fiber. Recent theoretical work, based on electron-microscopy images
of reconstituted fibers supports this view.
DNA loops
The beads-on-a-string chromatin structure has a tendency to form loops. These loops allow interactions between different regions of DNA by bringing them closer to each other, which increases the efficiency of gene interactions. This process is dynamic, with loops forming and disappearing. The loops are regulated by two main elements: *Cohesin
Cohesin is a protein complex that mediates Establishment of sister chromatid cohesion, sister chromatid cohesion, homologous recombination, and Topologically associating domain, DNA looping. Cohesin is formed of SMC3, SMC1A, SMC1, RAD21, SCC1 an ...
s, protein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple active site, catalytic domains are found in a single polypeptide chain.
...
es that generate loops by extrusion of the DNA fiber through the ring-like structure of the complex itself.
* CTCF
Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the ''CTCF'' gene. CTCF is involved in many cellular processes, including transcriptional regulati ...
, a transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
that limits the frontier of the DNA loop. To stop the growth of a loop, two CTCF molecules must be positioned in opposite directions to block the movement of the cohesin ring (''see video'').
There are many other elements involved. For example, Jpx regulates the binding sites of CTCF molecules along the DNA fiber.
Spatial organization of chromatin in the cell nucleus
The spatial arrangement of the chromatin within the nucleus is not random - specific regions of the chromatin can be found in certain territories. Territories are, for example, the lamina-associated domains (LADs), and thetopologically associating domain
A topologically associating domain (TAD) is a self-interacting genomic region, meaning that DNA sequences within a TAD physically interact with each other more frequently than with sequences outside the TAD. The average size of a topologically ass ...
s (TADs), which are bound together by protein complexes. Currently, polymer models such as the Strings & Binders Switch (SBS) model and the Dynamic Loop (DL) model are used to describe the folding of chromatin within the nucleus. The arrangement of chromatin within the nucleus may also play a role in nuclear stress and restoring nuclear membrane deformation by mechanical stress. When chromatin is condensed, the nucleus becomes more rigid. When chromatin is decondensed, the nucleus becomes more elastic with less force
In physics, a force is an influence that can cause an Physical object, object to change its velocity unless counterbalanced by other forces. In mechanics, force makes ideas like 'pushing' or 'pulling' mathematically precise. Because the Magnitu ...
exerted on the inner nuclear membrane. This observation sheds light on other possible cellular functions of chromatin organization outside of genomic regulation.
Cell-cycle dependent structural organization

interphase
Interphase is the active portion of the cell cycle that includes the G1, S, and G2 phases, where the cell grows, replicates its DNA, and prepares for mitosis, respectively. Interphase was formerly called the "resting phase," but the cell i ...
of mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
is optimized to allow simple access of transcription and DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
factors to the DNA while compacting the DNA into the nucleus
Nucleus (: nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucleu ...
. The structure varies depending on the access required to the DNA. Genes
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
that require regular access by RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
require the looser structure provided by euchromatin.
# Metaphase: The metaphase
Metaphase ( and ) is a stage of mitosis in the eukaryotic cell cycle in which chromosomes are at their second-most condensed and coiled stage (they are at their most condensed in anaphase). These chromosomes, carrying genetic information, alig ...
structure of chromatin differs vastly to that of interphase
Interphase is the active portion of the cell cycle that includes the G1, S, and G2 phases, where the cell grows, replicates its DNA, and prepares for mitosis, respectively. Interphase was formerly called the "resting phase," but the cell i ...
. It is optimised for physical strength and manageability, forming the classic chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
structure seen in karyotype
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by de ...
s. The structure of the condensed chromatin is thought to be loops of 30 nm fibre to a central scaffold of proteins. It is, however, not well-characterised. Chromosome scaffolds play an important role to hold the chromatin into compact chromosomes. Loops of 30 nm structure further condense with scaffold, into higher order structures. Chromosome scaffolds are made of proteins including condensin
Condensins are large protein complexes that play a central role in chromosome condensation and segregation during mitosis and meiosis (Figure 1). Their subunits were originally identified as major components of mitotic chromosomes assembled in ' ...
, type IIA topoisomerase and kinesin family member 4 (KIF4). The physical strength of chromatin is vital for this stage of division to prevent shear damage to the DNA as the daughter chromosomes are separated. To maximise strength the composition of the chromatin changes as it approaches the centromere, primarily through alternative histone H1 analogues. During mitosis, although most of the chromatin is tightly compacted, there are small regions that are not as tightly compacted. These regions often correspond to promoter regions of genes that were active in that cell type prior to chromatin formation. The lack of compaction of these regions is called bookmarking, which is an epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
mechanism believed to be important for transmitting to daughter cells the "memory" of which genes were active prior to entry into mitosis. This bookmarking mechanism is needed to help transmit this memory because transcription ceases during mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
.
Chromatin and bursts of transcription
Chromatin and its interaction with enzymes has been researched, and a conclusion being made is that it is relevant and an important factor in gene expression. Vincent G. Allfrey, a professor at Rockefeller University, stated that RNA synthesis is related to histone acetylation. The lysine amino acid attached to the end of the histones is positively charged. The acetylation of these tails would make the chromatin ends neutral, allowing for DNA access. When the chromatin decondenses, the DNA is open to entry of molecular machinery. Fluctuations between open and closed chromatin may contribute to the discontinuity of transcription, or transcriptional bursting. Other factors are probably involved, such as the association and dissociation of transcription factor complexes with chromatin. Specifically, RNA polymerase and transcriptional proteins have been shown to congregate into droplets via phase separation, and recent studies have suggested that 10 nm chromatin demonstrates liquid-like behavior increasing the targetability of genomic DNA. The interactions between linker histones and disordered tail regions act as an electrostatic glue organizing large-scale chromatin into a dynamic, liquid-like domain. Decreased chromatin compaction comes with increased chromatin mobility and easier transcriptional access to DNA. The phenomenon, as opposed to simple probabilistic models of transcription, can account for the high variability in gene expression occurring between cells in isogenic populations.Alternative chromatin organizations
During metazoanspermiogenesis
Spermiogenesis is the final stage of spermatogenesis, during which the spermatids develop into mature spermatozoon, spermatozoa. At the beginning of the stage, the spermatid is a more or less circular cell containing a cell nucleus, nucleus, Golg ...
, the spermatid
The spermatid is the haploid male gametid that results from division of secondary spermatocytes. As a result of meiosis, each spermatid contains only half of the genetic material present in the original primary spermatocyte.
Spermatids are co ...
's chromatin is remodeled into a more spaced-packaged, widened, almost crystal-like structure. This process is associated with the cessation of transcription and involves nuclear
Nuclear may refer to:
Physics
Relating to the nucleus of the atom:
*Nuclear engineering
*Nuclear physics
*Nuclear power
*Nuclear reactor
*Nuclear weapon
*Nuclear medicine
*Radiation therapy
*Nuclear warfare
Mathematics
* Nuclear space
*Nuclear ...
protein exchange. The histones are mostly displaced, and replaced by protamine
Protamines are small, arginine-rich, nuclear proteins that replace histones late in the haploid phase of spermatogenesis and are believed essential for sperm head condensation and DNA stabilization. They may allow for denser packaging of DNA ...
s (small, arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
-rich proteins). It is proposed that in yeast, regions devoid of histones become very fragile after transcription; HMO1, an HMG-box protein, helps in stabilizing nucleosomes-free chromatin.
Chromatin and DNA repair
A variety of internal and external agents can cause DNA damage in cells. Many factors influence how the repair route is selected, including the cell cycle phase and chromatin segment where the break occurred. In terms of initiating 5’ end DNA repair, the p53 binding protein 1 ( 53BP1) andBRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a ...
are important protein components that influence double-strand break repair pathway selection. The 53BP1 complex attaches to chromatin near DNA breaks and activates downstream factors such as Rap1-Interacting Factor 1 ( RIF1) and shieldin, which protects DNA ends against nucleolytic destruction. DNA damage process occurs within the condition of chromatin, and the constantly changing chromatin environment has a large effect on it. Accessing and repairing the damaged cell of DNA, the genome condenses into chromatin and repairing it through modifying the histone residues. Through altering the chromatin structure, histones residues are adding chemical groups namely phosphate, acetyl and one or more methyl groups and these control the expressions of gene building by proteins to acquire DNA. Moreover, resynthesis of the delighted zone, DNA will be repaired by processing and restructuring the damaged bases. In order to maintain genomic integrity, "homologous recombination and classical non-homologous end joining process" has been followed by DNA to be repaired.
The packaging of eukaryotic DNA into chromatin presents a barrier to all DNA-based processes that require recruitment of enzymes to their sites of action. To allow the critical cellular process of DNA repair, the chromatin must be remodeled. In eukaryotes, ATP-dependent chromatin remodeling complexes and histone-modifying enzymes are two predominant factors employed to accomplish this remodeling process.
Chromatin relaxation occurs rapidly at the site of DNA damage. This process is initiated by PARP1 protein that starts to appear at DNA damage in less than a second, with half maximum accumulation within 1.6 seconds after the damage occurs. Next the chromatin remodeler Alc1 quickly attaches to the product of PARP1, and completes arrival at the DNA damage within 10 seconds of the damage. About half of the maximum chromatin relaxation, presumably due to action of Alc1, occurs by 10 seconds. This then allows recruitment of the DNA repair enzyme MRE11, to initiate DNA repair, within 13 seconds.
γH2AX, the phosphorylated form of H2AX is also involved in the early steps leading to chromatin decondensation after DNA damage occurrence. The histone variant H2AX constitutes about 10% of the H2A histones in human chromatin. γH2AX (H2AX phosphorylated on serine 139) can be detected as soon as 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurs in one minute. The extent of chromatin with phosphorylated γH2AX is about two million base pairs at the site of a DNA double-strand break. γH2AX does not, itself, cause chromatin decondensation, but within 30 seconds of irradiation, RNF8
E3 ubiquitin-protein ligase RNF8 is an enzyme that in humans is encoded by the ''RNF8'' gene. RNF8 has activity both in immune system functions and in DNA repair.
Function
The protein encoded by this gene contains a RING finger domain, RING f ...
protein can be detected in association with γH2AX. RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4
Chromodomain-helicase-DNA-binding protein 4 is an enzyme that in humans is encoded by the ''CHD4'' gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleo ...
, a component of the nucleosome remodeling and deacetylase complex NuRD.
After undergoing relaxation subsequent to DNA damage, followed by DNA repair, chromatin recovers to a compaction state close to its pre-damage level after about 20 min.
Methods to investigate chromatin

 #
# ChIP-seq
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with Massively parallel signature sequencing, massively parallel DNA sequencing to identify t ...
(Chromatin immunoprecipitation sequencing) is recognized as the vastly utilized chromatin identification method it has been using the antibodies that actively selected, identify and combine with proteins including "histones, histone restructuring, transaction factors and cofactors". This has been providing data about the state of chromatin and the transaction of a gene by trimming "oligonucleotides" that are unbound. Chromatin immunoprecipitation sequencing aimed against different histone modification
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes. ...
s, can be used to identify chromatin states throughout the genome. Different modifications have been linked to various states of chromatin.
# DNase-seq DNase-seq ( DNase I hypersensitive sites sequencing) is a method in molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, i ...
(DNase I hypersensitive sites Sequencing) uses the sensitivity of accessible regions in the genome to the DNase I enzyme to map open or accessible regions in the genome.
# FAIRE-seq (Formaldehyde
Formaldehyde ( , ) (systematic name methanal) is an organic compound with the chemical formula and structure , more precisely . The compound is a pungent, colourless gas that polymerises spontaneously into paraformaldehyde. It is stored as ...
-Assisted Isolation of Regulatory Elements sequencing) uses the chemical properties of protein-bound DNA in a two-phase separation method to extract nucleosome depleted regions from the genome.
# ATAC-seq
ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) is a laboratory technique used in molecular biology to assess genome-wide chromatin, chromatin accessibility. The technique was first described in 2013 as an alternative approa ...
(Assay for Transposable Accessible Chromatin sequencing) uses the Tn5 transposase to integrate (synthetic) transposons into accessible regions of the genome consequentially highlighting the localisation of nucleosomes and transcription factors across the genome.
# DNA footprinting is a method aimed at identifying protein-bound DNA. It uses labeling and fragmentation coupled to gel electrophoresis to identify areas of the genome that have been bound by proteins.
# MNase-seq (Micrococcal Nuclease sequencing) uses the micrococcal nuclease enzyme to identify nucleosome positioning throughout the genome.
# Chromosome conformation capture determines the spatial organization of chromatin in the nucleus, by inferring genomic locations that physically interact.
# MACC profiling (Micrococcal nuclease ACCessibility profiling) uses titration series of chromatin digests with micrococcal nuclease to identify chromatin accessibility as well as to map nucleosomes and non-histone DNA-binding proteins in both open and closed regions of the genome.
Chromatin and knots
It has been a puzzle how decondensed interphase chromosomes remain essentially unknotted. The natural expectation is that in the presence of type II DNA topoisomerases that permit passages of double-stranded DNA regions through each other, all chromosomes should reach the state of topological equilibrium. The topological equilibrium in highly crowded interphase chromosomes forming chromosome territories would result in formation of highly knotted chromatin fibres. However, Chromosome Conformation Capture (3C) methods revealed that the decay of contacts with the genomic distance in interphase chromosomes is practically the same as in the crumpled globule state that is formed when long polymers condense without formation of any knots. To remove knots from highly crowded chromatin, one would need an active process that should not only provide the energy to move the system from the state of topological equilibrium but also guide topoisomerase-mediated passages in such a way that knots would be efficiently unknotted instead of making the knots even more complex. It has been shown that the process of chromatin-loop extrusion is ideally suited to actively unknot chromatin fibres in interphase chromosomes.Chromatin: alternative definitions
The term, introduced byWalther Flemming
Walther Flemming (21 April 1843 – 4 August 1905) was a German biologist and a founder of cytogenetics.
He was born in Sachsenberg (now part of Schwerin) as the fifth child and only son of the psychiatrist Carl Friedrich Flemming (1799–1880 ...
, has multiple meanings:
# Simple and concise definition: Chromatin is a macromolecular complex of a DNA macromolecule and protein macromolecules (and RNA). The proteins package and arrange the DNA and control its functions within the cell nucleus.
# A biochemists' operational definition: Chromatin is the DNA/protein/RNA complex extracted from eukaryotic lysed interphase nuclei. Just which of the multitudinous substances present in a nucleus will constitute a part of the extracted material partly depends on the technique each researcher uses. Furthermore, the composition and properties of chromatin vary from one cell type to another, during the development of a specific cell type, and at different stages in the cell cycle.
# The ''DNA + histone = chromatin'' definition: The DNA double helix in the cell nucleus is packaged by special proteins termed histones. The formed protein/DNA complex is called chromatin. The basic structural unit of chromatin is the nucleosome.
The first definition allows for "chromatins" to be defined in other domains of life like bacteria and archaea, using any DNA-binding proteins that condenses the molecule. These proteins are usually referred to nucleoid-associated proteins (NAPs); examples include AsnC/LrpC with HU. In addition, some archaea do produce nucleosomes from proteins homologous to eukaryotic histones.
Chromatin Remodeling:
Chromatin remodeling can result from covalent modification of histones that physically remodel, move or remove nucleosomes. Studies of Sanosaka et al. 2022, says that Chromatin remodeler CHD7 regulate cell type-specific gene expression in human neural crest cells.
See also
* Active chromatin sequence *Chromatid
A chromatid (Greek ''khrōmat-'' 'color' + ''-id'') is one half of a duplicated chromosome. Before replication, one chromosome is composed of one DNA molecule. In replication, the DNA molecule is copied, and the two molecules are known as chrom ...
* ENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims "to build a comprehensive parts list of functional elements in the human genome."
ENCODE also supports further biomedical research by "generating community resourc ...
database
* Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
* Histone-modifying enzymes
* Position-effect variegation Position-effect variegation (PEV) is a Variegation (histology), variegation caused by the silencing of a gene in some cells through its abnormal juxtaposition with heterochromatin via rearrangement or Transposable element, transposition. It is also ...
* Transcriptional bursting
Notes
References
Additional sources
* Cooper, Geoffrey M. 2000. The Cell, 2nd edition, A Molecular Approach. Chapter 4.Chromosomes and Chromatin.
* * Cremer, T. 1985. Von der Zellenlehre zur Chromosomentheorie: Naturwissenschaftliche Erkenntnis und Theorienwechsel in der frühen Zell- und Vererbungsforschung, Veröffentlichungen aus der Forschungsstelle für Theoretische Pathologie der Heidelberger Akademie der Wissenschaften. Springer-Vlg., Berlin, Heidelberg. * Elgin, S. C. R. (ed.). 1995. Chromatin Structure and Gene Expression, vol. 9. IRL Press, Oxford, New York, Tokyo. * * * * * * Pollard, T., and W. Earnshaw. 2002. Cell Biology. Saunders. * Saumweber, H. 1987. Arrangement of Chromosomes in Interphase Cell Nuclei, p. 223-234. In W. Hennig (ed.), Structure and Function of Eucaryotic Chromosomes, vol. 14. Springer-Verlag, Berlin, Heidelberg. * * Van Holde KE. 1989. Chromatin. New York:
Springer-Verlag
Springer Science+Business Media, commonly known as Springer, is a German multinational publishing company of books, e-books and peer-reviewed journals in science, humanities, technical and medical (STM) publishing.
Originally founded in 1842 in ...
. .
* Van Holde, K., J. Zlatanova, G. Arents, and E. Moudrianakis. 1995. Elements of chromatin structure: histones, nucleosomes, and fibres, p. 1-26. In S. C. R. Elgin (ed.), Chromatin structure and gene expression. IRL Press at Oxford University Press, Oxford.
External links
Chromatin, Histones & Cathepsin
PMAP The Proteolysis Map-animation
''Nature'' journal: recent chromatin publications and news
Protocol for ''in vitro'' Chromatin Assembly
ENCODE threads Explorer
Chromatin patterns at transcription factor binding sites.
Nature (journal)
''Nature'' is a British weekly scientific journal founded and based in London, England. As a multidisciplinary publication, ''Nature'' features Peer review, peer-reviewed research from a variety of academic disciplines, mainly in science and t ...
{{portal bar, Biology, Science
Molecular genetics
Nuclear substructures