|
MRE11A
Double-strand break repair protein MRE11 (Meiotic recombination 11) is an enzyme that in humans is encoded by the ''MRE11'' gene. The gene has been designated ''MRE11A'' to distinguish it from the pseudogene ''MRE11B'' that is nowadays named ''MRE11P1''. Function This gene encodes a nuclear protein involved in homologous recombination, telomere length maintenance, and DNA double-strand break repair. By itself, the protein has 3' to 5' exonuclease activity and endonuclease activity. The protein forms a complex with the RAD50 homolog; this complex is required for nonhomologous joining of DNA ends and possesses increased single-stranded DNA endonuclease and 3' to 5' exonuclease activities. In conjunction with a DNA ligase, this protein promotes the joining of noncomplementary ends in vitro using short homologies near the ends of the DNA fragments. This gene has a pseudogene on chromosome 3. Alternative splicing of this gene results in two transcript variants encoding different is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair, repair harmful DNA breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and ovum, egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to Adaptation, adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nibrin
Nibrin, also known as NBN or NBS1, is a protein which in humans is encoded by the ''NBN'' gene. Function Nibrin is a protein associated with the repair of double strand breaks (DSBs) which pose serious damage to a genome. It is a 754 amino acid protein identified as a member of the NBS1/hMre11/RAD50(N/M/R, more commonly referred to as MRN) double strand DNA break repair complex. This complex recognizes DNA damage and rapidly relocates to DSB sites and forms nuclear foci. It also has a role in regulation of N/M/R (MRN) protein complex activity which includes end-processing of both physiological and mutagenic DNA double strand breaks (DSBs). Cellular response to DSBs Cellular response is performed by damage sensors, effectors of lesion repair and signal transduction. The central role is carried out by ataxia telangiectasia mutated (ATM) by activating the DSB signaling cascade, phosphorylating downstream substrates such as histone H2AX and NBS1. NBS1 relocates to DSB sit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rad50
DNA repair protein RAD50, also known as RAD50, is a protein that in humans is encoded by the ''RAD50'' gene. Function The protein encoded by this gene is highly similar to ''Saccharomyces cerevisiae'' Rad50, a protein involved in DNA double-strand break repair. This protein forms a complex with MRE11 and NBS1 (also known as Xrs2 in yeast). This MRN complex (MRX complex in yeast) binds to broken DNA ends and displays numerous enzymatic activities that are required for double-strand break repair by nonhomologous end-joining or homologous recombination. Gene knockout studies of the mouse homolog of Rad50 suggest it is essential for cell growth and viability. Two alternatively spliced transcript variants of Rad50, which encode distinct proteins, have been reported. Structure Rad50 is a member of the structural maintenance of chromosomes (SMC) family of proteins. Like other SMC proteins, Rad50 contains a long internal coiled-coil domain that folds back on itself, bringing t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair-deficiency Disorder
A DNA repair-deficiency disorder is a medical condition due to reduced functionality of DNA repair. DNA repair defects can cause an accelerated aging disease or an increased risk of cancer, or sometimes both. DNA repair defects and accelerated aging DNA repair defects are seen in nearly all of the diseases described as accelerated aging disease, in which various tissues, organs or systems of the human body age prematurely. Because the accelerated aging diseases display different aspects of aging, but never every aspect, they are often called segmental progerias by biogerontologists. Human disorders with accelerated aging * Ataxia-telangiectasia * Bloom syndrome * Cockayne syndrome * Fanconi anemia * Progeria (Hutchinson–Gilford progeria syndrome) * Rothmund–Thomson syndrome * Trichothiodystrophy * Werner syndrome * Xeroderma pigmentosum Examples Some examples of DNA repair defects causing progeroid syndromes in humans or mice are shown in Table 1. DNA repair ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer Syndrome
A hereditary cancer syndrome (familial/family cancer syndrome, inherited cancer syndrome, cancer predisposition syndrome, cancer syndrome, etc.) is a genetic disorder in which inherited genetic mutations in one or more genes predispose the affected individuals to the development of cancer and may also cause early onset of these cancers. Hereditary cancer syndromes often show not only a high lifetime risk of developing cancer, but also the development of multiple independent primary tumors. Many of these syndromes are caused by mutations in tumor suppressor genes, genes that are involved in protecting the cell from turning cancerous. Other genes that may be affected are DNA repair genes, oncogenes and genes involved in the production of blood vessels (angiogenesis). Common examples of inherited cancer syndromes are hereditary breast-ovarian cancer syndrome and hereditary non-polyposis colon cancer (Lynch syndrome). Background Hereditary cancer syndromes underlie 5 to 10% of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TERF2
Telomeric repeat-binding factor 2 is a protein that is present at telomeres throughout the cell cycle. It is also known as TERF2, TRF2, and TRBF2, and is encoded in humans by the ''TERF2'' gene. It is a component of the shelterin nucleoprotein complex and a second negative regulator of telomere length, playing a key role in the protective activity of telomeres. It was first reported in 1997 in the lab of Titia de Lange, where a DNA sequence similar, but not identical, to TERF1 was discovered, with respect to the Myb-domain. De Lange isolated the new Myb-containing protein sequence and called it TERF2. Structure and domains TERF2 has a similar structure to that of TERF1. Both proteins carry a C-terminus Myb motif and large TERF1-related dimerization domains near their N-terminus. However, both proteins exist exclusively as homodimers and do not heterodimerize with each other, as proven by co-immunoprecipitation assay analysis. Also, TERF2 has a basic N-terminus, differing fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Double-strand Break
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in cells, by internal metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair processes fail, including apoptosis, irreparable DNA damage may occur, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MDC1
Mediator of DNA damage checkpoint protein 1 is a 2080 amino acid long protein that in humans is encoded by the ''MDC1'' gene located on the short arm (p) of chromosome 6. MDC1 protein is a regulator of the Intra-S phase and the G2/M cell cycle checkpoints and recruits repair proteins to the site of DNA damage. It is involved in determining cell survival fate in association with tumor suppressor protein p53. This protein also goes by the name Nuclear Factor with BRCT Domain 1 (NFBD1). Function Role in DNA damage response The ''MDC1'' gene encodes the MDC1 nuclear protein which is part of the DNA damage response (DDR) pathway, the mechanism through which eukaryotic cells respond to damaged DNA, specifically DNA double-strand breaks (DSB) that are caused by ionizing radiation or chemical clastogens. The DDR of mammalian cells is made up of kinases, and mediator/adaptors factors. In mammalian cells the DDR is a network of pathways made up of proteins that function as either ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ku70
Ku70 is a heterodimeric protein made up of Ku70 and Ku80, which together form Ku. In humans, is encoded by the ''XRCC6'' gene. Ku70 plays a critical role in the DNA repair, maintenance and many other cellular processes. Function Together, Ku70 and Ku80 make up the Ku heterodimer form a quasi-symmetric structure, which encircles the double-stranded DNA. The DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) of the DNA repair pathway. It is also required for V(D)J recombination, which utilizes the NHEJ pathway to promote antigen diversity in the mammalian immune system. Ku70 is key for sensing and responding to cytosolic DNA, which is essential for the indication of infection. Within the heterodimer, Ku70 specifically binds directly to broken ends of double-stranded DNA breaks, or DSBs. Then together, Ku70 and Ku80 will tightly form a ring-like structure around the DNA strand, preventing further degradation. These steps are essential for the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
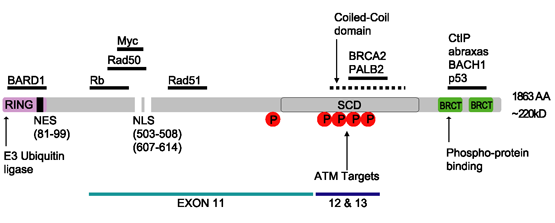
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a human tumor suppressor gene (also known as a caretaker gene) and is responsible for repairing DNA. ''BRCA1'' and ''BRCA2'' are unrelated proteins, but both are normally expressed in the cells of breast and other tissues, where they help repair damaged DNA, or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks. If ''BRCA1'' or ''BRCA2'' itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer. ''BRCA1'' and ''BRCA2'' have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a no ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
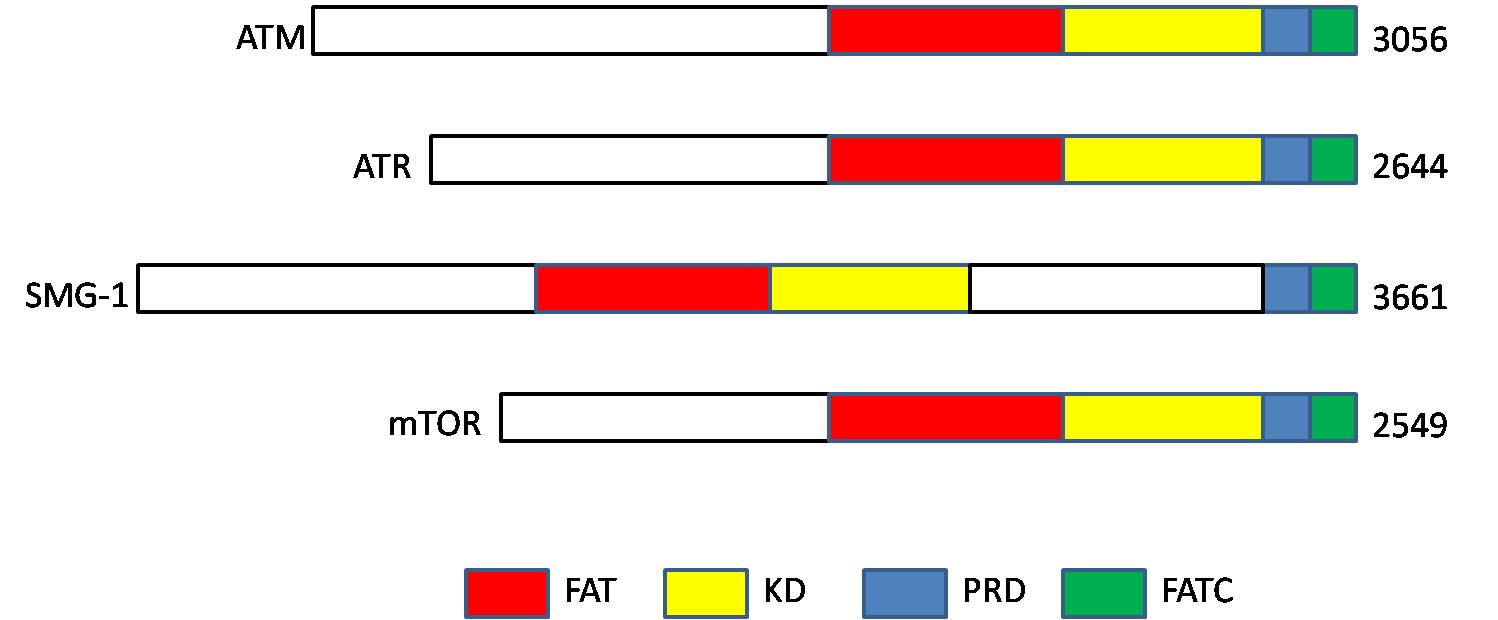
Ataxia Telangiectasia Mutated
ATM serine/threonine kinase or Ataxia-telangiectasia mutated, symbol ATM, is a serine/threonine protein kinase that is recruited and activated by DNA repair#Double-strand breaks, DNA double-strand breaks (Canonical pathway, canonical pathway), oxidative stress, topoisomerase cleavage complexes, splicing intermediates, R-loops and in some cases by DNA repair, single-strand DNA breaks. It phosphorylates several key proteins that initiate activation of the DNA damage cell cycle checkpoint, checkpoint, leading to cell cycle arrest, DNA repair or apoptosis. Several of these targets, including p53, CHK2, BRCA1, NBS1 and H2AX are tumor suppressors. In 1995, the gene was discovered by Yosef Shiloh who named its product ATM since he found that its mutations are responsible for the disorder ataxia–telangiectasia#Cause, ataxia–telangiectasia. In 1998, the Shiloh and Michael B. Kastan, Kastan laboratories independently showed that ATM is a protein kinase whose activity is enhanced by DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |