|
HMG-box
In molecular biology, the HMG-box (high mobility group box) is a protein domain which is involved in DNA binding. The domain is composed of approximately 75 amino acid residues that collectively mediate the DNA-binding of chromatin-associated high-mobility group proteins. HMG-boxes are present in many transcription factors and chromatin-remodeling complexes, where they can mediate non-sequence or sequence-specific DNA binding. Structure The structure of the HMG-box domain contains three alpha helices separated by loops (see figure to the right). Function HMG-box containing proteins only bind non- B-type DNA conformations (kinked or unwound) with high affinity. HMG-box domains are found in some high mobility group proteins, which are involved in the regulation of DNA-dependent processes such as transcription, replication, and DNA repair DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High Mobility Group
High-Mobility Group or HMG is a group of chromosomal proteins that are involved in the regulation of DNA-dependent processes such as transcription, replication, recombination, and DNA repair. History and name HMG proteins were originally isolated from mammalian cells, and named according to their electrophoretic mobility in polyacrylamide gels. Families The HMG proteins are subdivided into 3 superfamilies each containing a characteristic functional domain: * HMGA – contains an AT-hook domain ** HMGA1 ** HMGA2 * HMGB – contains a HMG-box domain ** HMGB1 ** HMGB2 ** HMGB3 ** HMGB4 * HMGN – contains a nucleosomal binding domain ** HMGN1 ** HMGN2 ** HMGN3 ** HMGN4 ** HMGN5 Proteins containing any of the above domains embedded in their sequence are known as HMG-motif proteins. HMG-box proteins are found in a variety of eukaryotic organisms. Other families with HMG-box domain * SOX gene family ** Sex-Determining Region Y Protein ** SOX1, SOX2, etc. * TCF/ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
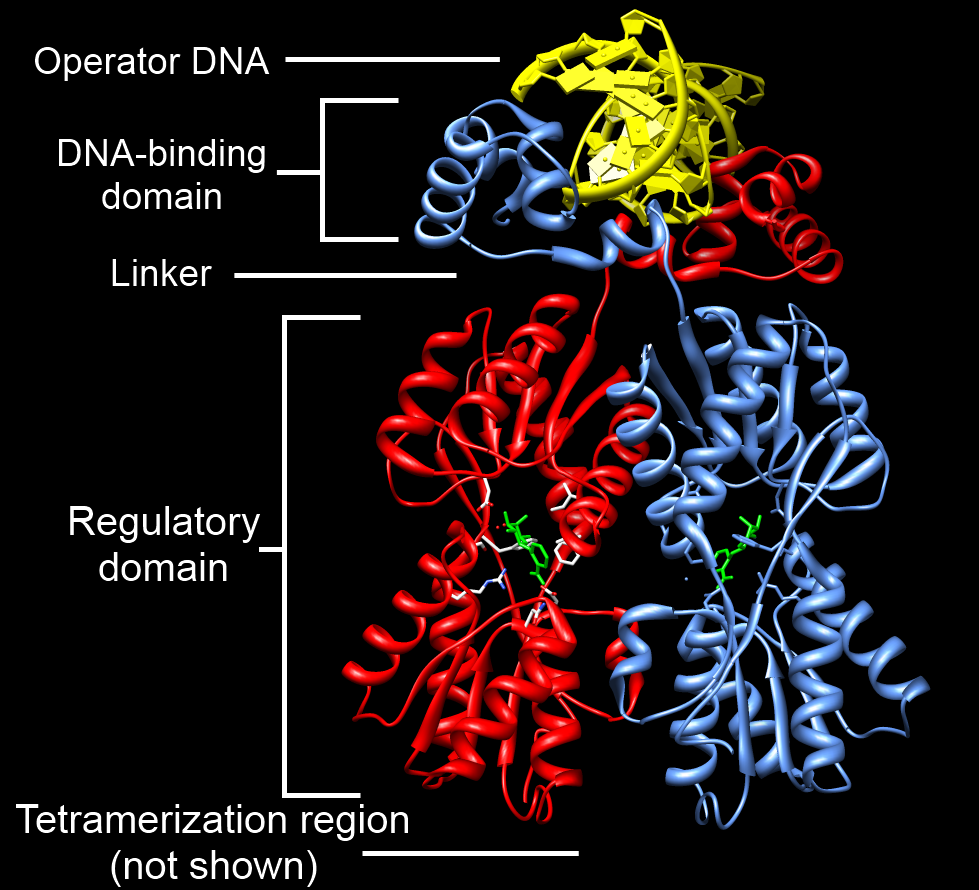
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA repair#DNA damage, DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30 nm fiber. ''Current opinion in cell biolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Nuclear Magnetic Resonance Spectroscopy
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conformational Isomerism
In chemistry, rotamers are chemical species that differ from one another primarily due to rotations about one or more single bonds. Various arrangements of atoms in a molecule that differ by rotation about single bonds can also be referred to as conformations. Conformers/rotamers differ little in their energies, so they are almost never separable in a practical sense. Rotations about single bonds are subject to small energy barriers. When the time scale for interconversion is long enough for isolation of individual rotamers (usually arbitrarily defined as a half-life of interconversion of 1000 seconds or longer), the species are termed atropisomers (''see:'' atropisomerism). The Ring flip, ring-flip of substituted cyclohexanes constitutes a common form of conformers. The study of the energetics of bond rotation is referred to as conformational analysis. In some cases, conformational analysis can be used to predict and explain product selectivity, mechanisms, and rates of reaction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a nucleic acid double helix, double helix of two Complementary DNA, complementary DNA strand, strands. DNA is often called double helix. The double helix describes the appearance of a double-stranded DNA which is composed of two linear strands that run opposite to each other and twist together. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (genetics)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs). Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. In virology, the term transcription is used when referring to mRNA synthesis from a viral RNA molecule. The genome of many RNA viruses is composed of negative-sense RNA which acts as a template for positive sense viral messenger RNA - a necessary step in the synthesis of viral proteins needed for viral replication. This process is catalyzed by a viral RNA dependent RNA polymerase. Background A DNA transcription unit encoding ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
B-DNA
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its tertiary structure. The structure was discovered by Rosalind Franklin and her student Raymond Gosling, Maurice Wilkins, James Watson, and Francis Crick, while the term "double helix" entered popular culture with the 1968 publication of Watson's '' The Double Helix: A Personal Account of the Discovery of the Structure of DNA''. The DNA double helix biopolymer of nucleic acid is held together by nucleotides which base pair together. In B-DNA, the most common double helical structure found in nature, the double helix is right-handed with about 10–10.5 base pairs per turn. The double helix structure of DNA contains a ''major groove'' and ''minor groove''. In B-DNA the major groove is wider th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lymphoid Enhancer-binding Factor 1
Lymphoid enhancer-binding factor 1 (LEF1) is a protein that in humans is encoded by the ''LEF1'' gene. It is a member of T cell factor/lymphoid enhancer factor (TCF/LEF family, TCF/LEF) family. Function Lymphoid enhancer-binding factor-1 (LEF1) is a 48-kD nuclear protein that is expressed in pre-B cell, B and T cells. It binds to a functionally important site in the T-cell receptor-alpha (TRA@, TCRA) enhancer and confers maximal enhancer activity. LEF1 belongs to a family of regulatory proteins that share homology with high mobility group protein-1 (HMGB1, HMG1). These High-mobility group, high mobility groups regulate a vast array of cellular processes through the production of transcription factors, which go on to regulate some of the cells most vital processes, including chromatin remodeling, Recombination signal sequences, recombination, DNA replication, DNA repair and transcription. In this shared homology between LEF1 and other members of the HMG family, many of the mech ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |