|
Histone Acetylation And Deacetylation
Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminus, N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation. Histone acetylation and deacetylation are essential parts of Regulation of gene expression, gene regulation. These reactions are typically catalysed by enzymes with "histone acetyltransferase" (HAT) or "histone deacetylase" (HDAC) activity. Acetylation is the process where an acetyl functional group is transferred from one molecule (in this case, acetyl coenzyme A) to another. Deacetylation is simply the reverse reaction where an acetyl group is removed from a molecule. Acetylated histones, octameric proteins that organize chromatin into nucleosomes, the basic structural unit of the chromosomes and ultimately higher order structures, represent a type of epigenetic marker within chromatin. Acetylation removes the positive charge on the histones, thereb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
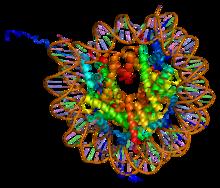
Nucleosome 1KX5 Colour Coded
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins Histone H2A, H2A, Histone H2B, H2B, Histone H3, H3, and Histone H4, H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry Epigenetics, epigenetically inherited information in the form of coval ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
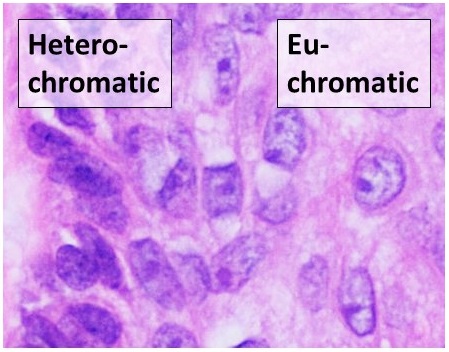
Euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic. In eukaryotes, euchromatin comprises the most active portion of the genome within the cell nucleus. In prokaryotes, euchromatin is the ''only'' form of chromatin present; this indicates that the heterochromatin structure evolved later along with the nucleus, possibly as a mechanism to handle increasing genome size. Structure Euchromatin is composed of repeating subunits known as nucleosomes, reminiscent of an unfolded set of beads on a string, that are approximately 11 nm in diameter. At the core of these nucleosomes are a set of four histone protein pairs: H3, H4, H2A, and H2B. Each core histone protein possesses a 'tail' structu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HAT1
Histone acetyltransferase 1, also known as HAT1, is an enzyme that, in humans, is encoded by the ''HAT1'' gene. Function The protein encoded by this gene is a type B histone acetyltransferase (HAT) that is involved in the rapid acetylation of newly synthesized cytoplasmic histones, which are, in turn, imported into the nucleus for de novo deposition onto nascent DNA chains. Histone acetylation, in particular, of histone H4 Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleo ..., plays an important role in replication-dependent chromatin assembly. To be specific, this HAT can acetylate soluble but not nucleosomal histone H4 at lysines 5 and 12, and, to a lesser degree, histone H2A at lysine 5. References Further reading * * * * * * * * * * * * * {{NLM content ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ELP3
Elongator complex protein 3, also named KAT9, is a protein that in humans is encoded by the ''ELP3'' gene. ELP3 is the catalytic histone acetyltransferase subunit of the RNA polymerase II elongator complex, which is a component of the RNA polymerase II (Pol II) holoenzyme and is involved in transcriptional elongation. ELP3 supports the migration and branching of projection neurons through acetylation of alpha-tubulin in the developing cerebral cortex. In mammals, ELP3 is important for paternal DNA demethylation after fertilization. ELP3 is potentially involved in cellular redox homeostasis by mediating the acetylation of glucose-6-phosphate dehydrogenase. Besides, ELP3 may play a role in chromatin remodeling Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out ... and is involved in acety ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PCAF
P300/CBP-associated factor (PCAF), also known as K(lysine) acetyltransferase 2B (KAT2B), is a human gene and transcriptional coactivator associated with p53. Structure Several domains of PCAF can act independently or in unison to enable its functions. PCAF has separate acetyltransferase and E3 ubiquitin ligase domains as well as a bromodomain for interaction with other proteins. PCAF also possesses sites for its own acetylation and ubiquitination. Function CBP and p300 are large nuclear proteins that bind to many sequence-specific factors involved in cell growth and/or differentiation, including c-jun and the adenoviral oncoprotein E1A. The protein encoded by the PCAF gene associates with p300/CBP. It has in vitro and in vivo binding activity with CBP and p300, and competes with E1A for binding sites in p300/CBP. It has histone acetyl transferase activity with core histones and nucleosome core particles, indicating that this protein plays a direct role in transcriptiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetylation And Deacetylation
Histone acetylation and deacetylation are the processes by which the lysine residues within the N-terminus, N-terminal tail protruding from the histone core of the nucleosome are acetylated and deacetylated as part of gene regulation. Histone acetylation and deacetylation are essential parts of Regulation of gene expression, gene regulation. These reactions are typically catalysed by enzymes with "histone acetyltransferase" (HAT) or "histone deacetylase" (HDAC) activity. Acetylation is the process where an acetyl functional group is transferred from one molecule (in this case, acetyl coenzyme A) to another. Deacetylation is simply the reverse reaction where an acetyl group is removed from a molecule. Acetylated histones, octameric proteins that organize chromatin into nucleosomes, the basic structural unit of the chromosomes and ultimately higher order structures, represent a type of epigenetic marker within chromatin. Acetylation removes the positive charge on the histones, thereb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetyl-coenzyme A
Acetyl-CoA (acetyl coenzyme A) is a molecule that participates in many biochemical reactions in protein, carbohydrate and lipid metabolism. Its main function is to deliver the acetyl group to the citric acid cycle (Krebs cycle) to be oxidized for energy production. Coenzyme A (CoASH or CoA) consists of a β-mercaptoethylamine group linked to pantothenic acid (vitamin B5) through an amide linkage and 3'-phosphorylated ADP. The acetyl group (indicated in blue in the structural diagram on the right) of acetyl-CoA is linked to the sulfhydryl substituent of the β-mercaptoethylamine group. This thioester linkage is a "high energy" bond, which is particularly reactive. Hydrolysis of the thioester bond is exergonic (−31.5 kJ/mol). CoA is acetylated to acetyl-CoA by the breakdown of carbohydrates through glycolysis and by the breakdown of fatty acids through β-oxidation. Acetyl-CoA then enters the citric acid cycle, where the acetyl group is oxidized to carbon dioxide and wa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Double Helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its tertiary structure. The structure was discovered by Rosalind Franklin and her student Raymond Gosling, Maurice Wilkins, James Watson, and Francis Crick, while the term "double helix" entered popular culture with the 1968 publication of Watson's '' The Double Helix: A Personal Account of the Discovery of the Structure of DNA''. The DNA double helix biopolymer of nucleic acid is held together by nucleotides which base pair together. In B-DNA, the most common double helical structure found in nature, the double helix is right-handed with about 10–10.5 base pairs per turn. The double helix structure of DNA contains a ''major groove'' and ''minor groove''. In B-DNA the major groove is wid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H4
Histone H4 is one of the five main histone proteins involved in the structure of chromatin in eukaryote, eukaryotic cells. Featuring a main globular domain and a long N-terminus, N-terminal tail, H4 is involved with the structure of the nucleosome of the 'beads on a string' organization. Histone proteins are highly post-translationally modified. Covalently bonded modifications include acetylation and methylation of the N-terminal tails. These modifications may alter Gene expression, expression of genes located on DNA associated with its parent histone octamer. Histone H4 is an important protein in the structure and function of chromatin, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Genetics Histone H4 is encoded in multiple genes at different loci including: HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'beads on a string' structure. Histone proteins are highly post-translationally modified however Histone H3 is the most extensively modified of the five histones. The term "Histone H3" alone is purposely ambiguous in that it does not distinguish between sequence variants or modification state. Histone H3 is an important protein in the emerging field of epigenetics, where its sequence variants and variable modification states are thought to play a role in the dynamic and long term regulation of genes. Epigenetics and post-translational modifications The N-terminus of H3 protrudes from the globular nucleosome core and is susceptible to post-translational modification that influence cellular processes. These modifications include the covalen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2B
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes. Structure Histone H2B is a lightweight structural protein made of 126 amino acids. Many of these amino acids have a positive charge at cellular pH, which allows them to interact with the negatively charged phosphate groups in DNA. Along with a central globular domain, histone H2B has two flexible histone tails that extend outwards – one at the N-terminal end and one at C-terminal end. These are highly involved in condensing chromatin from the beads-on-a-string conformation to a 30-nm fiber. Similar to other histone proteins, histone H2B has a distinct histone fold that is optimized for histone-histone as well as histone-DNA interactions. Two copies of histone H2B come together with two copies each of histone H2A, histone H3, and histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone H2A
Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells. The other histone proteins are: Histone H1, H1, Histone H2B, H2B, Histone H3, H3 and Histone H4, H4. Background Histones are proteins that package DNA into nucleosomes. Histones are responsible for maintaining the shape and structure of a nucleosome. One chromatin molecule is composed of at least one of each core histones per 100 base pairs of DNA. There are five families of histones known to date; these histones are termed H1/H5, H2A, H2B, H3, and H4. H2A is considered a core histone, along with H2B, H3 and H4. Core formation first occurs through the interaction of two H2A molecules. Then, H2A forms a Protein dimer, dimer with H2B; the core molecule is complete when H3-H4 also attaches to form a tetramer. Sequence variants Histone H2A is composed of non-allelic variants. The term "Histone H2A" is intentionally non-specific and refers to a variety of closely relat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |