|
A-DNA
A-DNA is one of the possible double helical structures which DNA can adopt. A-DNA is thought to be one of three biologically active double helical structures along with B-DNA and Z-DNA. It is a right-handed double helix fairly similar to the more common B-DNA form, but with a shorter, more compact helical structure whose base pairs are not perpendicular to the helix-axis as in B-DNA. It was discovered by Rosalind Franklin, who also named the A and B forms. She showed that DNA is driven into the A form when under dehydrating conditions. Such conditions are commonly used to form crystals, and many DNA crystal structures are in the A form. The same helical conformation occurs in double-stranded RNAs, and in DNA-RNA hybrid double helices. Structure Like the more common B-DNA, A-DNA is a right-handed double helix with major and minor grooves. However, as shown in the comparison table below, there is a slight increase in the number of base pairs (bp) per turn. This results in a smal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rosalind Franklin
Rosalind Elsie Franklin (25 July 192016 April 1958) was a British chemist and X-ray crystallographer. Her work was central to the understanding of the molecular structures of DNA (deoxyribonucleic acid), RNA (ribonucleic acid), viruses, coal, and graphite. Although her works on coal and viruses were appreciated in her lifetime, Franklin's contributions to the discovery of the structure of DNA were largely unrecognised during her life, for which Franklin has been variously referred to as the "wronged heroine", the "dark lady of DNA", the "forgotten heroine", a "feminist icon", and the "Sylvia Plath of molecular biology". Franklin graduated in 1941 with a degree in Natural Sciences (Cambridge), natural sciences from Newnham College, Cambridge, and then enrolled for a PhD in physical chemistry under Ronald George Wreyford Norrish, the 1920 Chair of Physical Chemistry at the University of Cambridge. Disappointed by Norrish's lack of enthusiasm, she took up a research position und ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
B-DNA
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, and is a fundamental component in determining its tertiary structure. The structure was discovered by Rosalind Franklin and her student Raymond Gosling, Maurice Wilkins, James Watson, and Francis Crick, while the term "double helix" entered popular culture with the 1968 publication of Watson's '' The Double Helix: A Personal Account of the Discovery of the Structure of DNA''. The DNA double helix biopolymer of nucleic acid is held together by nucleotides which base pair together. In B-DNA, the most common double helical structure found in nature, the double helix is right-handed with about 10–10.5 base pairs per turn. The double helix structure of DNA contains a ''major groove'' and ''minor groove''. In B-DNA the major groove is wider th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Tertiary Structure
Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized. Helical structures Double helix The double helix is the dominant tertiary structure for biological DNA, and is also a possible structure for RNA. Three DNA conformations are believed to be found in nature, A-DNA, B-DNA, and Z-DNA. The "B" form described by James D. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought to be one of three biologically active double-helical structures along with A-DNA and B-DNA. History Left-handed DNA was first proposed by Robert Wells and colleagues, as the structure of a repeating polymer of inosine–cytosine. They observed a "reverse" circular dichroism spectrum for such DNAs, and interpreted this incorrectly to mean that the strands wrapped around one another in a left-handed fashion. The relationship between Z-DNA and the more familiar B-DNA was indicated by the work of Pohl and Jovin, who showed that the ultraviolet circular dichroism of poly(dG-dC) was nearly inverted in 4 M sodium chloride solution and that the structure of poly d(I–C)·poly d(I–C) was in fact a right-handed D-DNA conformation. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-DNA
C-DNA, also known as C-form DNA, is one of many possible double helical conformations of DNA. DNA can be induced to take this form in particular conditions such as relatively low humidity and the presence of certain ions, such as Li+ or Mg2+, but C-form DNA is not very stable and does not occur naturally in living organisms. In 1961, it was found by Marvin, when he tried to repeat for the Li salt the higher water content pattern of the Na salt. What Marvin found is the semicrystalline C-DNA. "Semicrystalline" describes a diffraction pattern for which crystalline reflexions are seen at low resolution but continuous transform at higher resolution. Structure The C-DNA is a non-integral helix of slightly variable dimensions, with mean values of 3.32Å for the unit rise and 38.60° for the unit twist, giving about 9 1/3 rather that 10 unites per turn. There are some different models for C-DNA proposed over years. In 2000, van Dam and Levitt found that both C-DNA and B-DNA consist of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
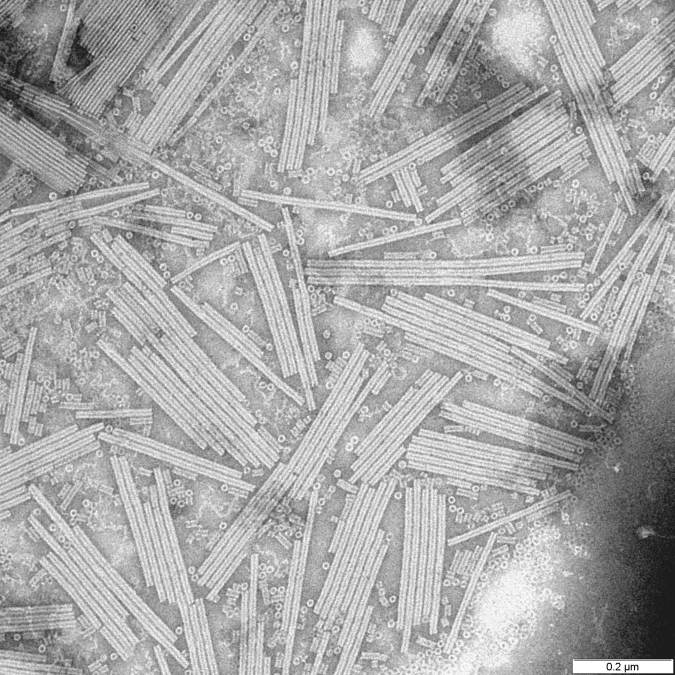
Sulfolobus Islandicus Filamentous Virus
''Betalipothrixvirus hveragerdiense'' (SIFV) is an archaeal virus, classified in the family ''Lipothrixviridae'' within the order ''Ligamenvirales''. The virus infects hyperthermophilic and acidophilic archaeon '' Sulfolobus islandicus''. SIFV has a linear double-stranded DNA genome of 40,852 bp, which is the largest among characterized lipothrixviruses. The virions are enveloped filaments, nearly 2 micrometers in length. The nucleocapsid is formed from two paralogous major capsid proteins, which tightly wrap around the dsDNA genome; notably, dehydration of the genomic DNA by the major capsid proteins transforms the B-form DNA into A-form DNA. Life cycle SIFV virions assemble inside the cell. Binding of the major capsid protein dimers to the linear dsDNA genome lead to the assembly of nucleocapsids, which are subsequently enveloped intracellularly through an unknown mechanism. SIFV and probably other lipothrixviruses are lytic viruses. Virion release takes place through pyram ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Förster Resonance Energy Transfer
Förster resonance energy transfer (FRET), fluorescence resonance energy transfer, resonance energy transfer (RET) or electronic energy transfer (EET) is a mechanism describing energy transfer between two light-sensitive molecules (chromophores). A donor chromophore, initially in its electronic excited state, may transfer energy to an acceptor chromophore through nonradiative dipole–dipole coupling. The efficiency of this energy transfer is inversely proportional to the sixth power of the distance between donor and acceptor, making FRET extremely sensitive to small changes in distance. Measurements of FRET efficiency can be used to determine if two fluorophores are within a certain distance of each other. Such measurements are used as a research tool in fields including biology and chemistry. FRET is analogous to Near and far field, near-field communication, in that the radius of interaction is much smaller than the Light, wavelength of light emitted. In the near-field region, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Portogloboviridae
''Portogloboviridae'' is a family of dsDNA viruses that infect archaea. It is a proposed family of the realm ''Varidnaviria'', but ICTV officially puts it as incertae sedis virus. Viruses in the family are related to '' Helvetiavirae''. The capsid proteins of these viruses and their characteristics are of evolutionary importance for the origin of the other ''Varidnaviria'' viruses since they seem to retain primordial characters. Description The virions in this family have a capsid with icosahedral geometry and a viral envelope that protects the genetic material. The diameter is 83 to 87 nanometers. The genome is circular dsDNA with a length of 20,222 base pairs. The genome contains 45 open reading frames (ORFs), which are closely arranged and occupy 89.1% of the genome. ORFs are generally short, with an average length of 103 codons. Virions have 10 proteins ranging from 20 to 32 kDa. Of these proteins, 8 code for the capsid and two for the viral envelope, including one that is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tristromaviridae
''Tristromaviridae'' is a family of viruses. Archaea Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ... of the genera '' Thermoproteus'' and '' Pyrobaculum'' serve as natural hosts. ''Tristromaviridae'' is the sole family in the order ''Primavirales''. There are two genera and three species in the family. Taxonomy The following genera and species are assigned to the family: * '' Alphatristromavirus'' ** '' Alphatristromavirus pozzuoliense'' ** '' Alphatristromavirus puteoliense'' * '' Betatristromavirus'' ** '' Betatristromavirus kraflaense'' Structure Viruses in the genus ''Tristromaviridae'' are enveloped, with rod-shaped geometries. The diameter is around 38 nm, with a length of 410 nm. Genomes are linear, around 15.9kb in length. The TTV1 virion contains four virus- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lipothrixviridae
''Lipothrixviridae'' is a family of viruses in the order '' Ligamenvirales''. Thermophilic archaea in the phylum Thermoproteota The Thermoproteota are prokaryotes that have been classified as a phylum (biology), phylum of the domain Archaea. Initially, the Thermoproteota were thought to be sulfur-dependent extremophiles but recent studies have identified characteristic T ... serve as natural hosts.Janekovic, D., Wunderl S, Holz I, Zillig W, Gierl A, Neumann H (1983) TTV1, TTV2 and TTV3, a family of viruses of the extremely thermophilic anaerobic, sulphur reducing, archaeabacterium Thermoproteus tenax. Mol. Gen. Genet. 19239–19245 Taxonomy The following genera and species are assigned to the family: * '' Alphalipothrixvirus'' * '' Betalipothrixvirus'' * '' Deltalipothrixvirus'' The family consists of three genera: ''Alphalipothrixvirus'', ''Betalipothrixvirus'', and ''Deltalipothrixvirus''. '' Captovirus'' used to be in this family as the genus Gammalipothrixvirus, but now ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |