Gene expression is the process (including its
regulation) by which information from a
gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
is used in the synthesis of a functional
gene product
A gene product is the biochemical material, either RNA or protein, resulting from the expression of a gene. A measurement of the amount of gene product is sometimes used to infer how active a gene is. Abnormal amounts of gene product can be corre ...
that enables it to produce end products,
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s or
non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally imp ...
, and ultimately affect a
phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
. These products are often
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s, but in non-protein-coding genes such as
transfer RNA (tRNA) and
small nuclear RNA (snRNA), the product is a functional
non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally imp ...
.
The process of gene expression is used by all known life—
eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
(including
multicellular organisms
A multicellular organism is an organism that consists of more than one cell, unlike unicellular organisms. All species of animals, land plants and most fungi are multicellular, as are many algae, whereas a few organisms are partially uni- and pa ...
),
prokaryotes
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
(
bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
and
archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
), and
virus
A virus is a submicroscopic infectious agent that replicates only inside the living Cell (biology), cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are ...
es—to generate the
macromolecular machinery for life.
In
genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian ...
, gene expression is the most fundamental level at which the
genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a ...
gives rise to the
phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
, ''i.e.'' observable trait. The genetic information stored in
DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
represents the genotype, whereas the phenotype results from the "interpretation" of that information. Such phenotypes are often displayed by the synthesis of proteins that control the organism's structure and development, or that act as
enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s catalyzing specific metabolic pathways.
All steps in the gene expression process may be modulated (regulated), including the
transcription,
RNA splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-cod ...
,
translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
, and
post-translational modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translation (biolog ...
of a protein.
Regulation of gene expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
gives control over the timing, location, and amount of a given gene product (protein or ncRNA) present in a cell and can have a profound effect on the cellular structure and function. Regulation of gene expression is the basis for
cellular differentiation
Cellular differentiation is the process in which a stem cell changes from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellula ...
,
development
Development or developing may refer to:
Arts
*Development (music), the process by which thematic material is reshaped
* Photographic development
*Filmmaking, development phase, including finance and budgeting
* Development hell, when a proje ...
,
morphogenesis
Morphogenesis (from the Greek ''morphê'' shape and ''genesis'' creation, literally "the generation of form") is the biological process that causes a cell, tissue or organism to develop its shape. It is one of three fundamental aspects of deve ...
and the versatility and
adaptability
Adaptability ( "fit to, adjust") is a feature of a system or of a process. This word has been put to use as a specialised term in different disciplines and in business operations. Word definitions of adaptability as a specialised term differ littl ...
of any
organism
An organism is any life, living thing that functions as an individual. Such a definition raises more problems than it solves, not least because the concept of an individual is also difficult. Many criteria, few of them widely accepted, have be ...
. Gene regulation may therefore serve as a substrate for evolutionary change.
Mechanism
Transcription
The production of a RNA copy from a DNA strand is called
transcription, and is performed by
RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
s, which add one
ribonucleotide at a time to a growing RNA strand as per the
complementarity law of the nucleotide bases. This RNA is
complementary to the template 3′ → 5′ DNA strand,
with the exception that
thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
s (T) are replaced with
uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
s (U) in the RNA and possible errors.
In bacteria, transcription is carried out by a single type of RNA polymerase, which needs to bind a DNA sequence called a
Pribnow box
The Pribnow box (also known as the Pribnow-Schaller box) is a sequence of ''TATAAT'' of six nucleotides (thymine, adenine, thymine, etc.) that is an essential part of a promoter site on DNA for transcription to occur in bacteria.
It is an ideal ...
with the help of the
sigma factor
A sigma factor (σ factor or specificity factor) is a protein needed for initiation of Transcription (biology), transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to g ...
protein (σ factor) to start transcription. In eukaryotes, transcription is performed in the nucleus by three types of RNA polymerases, each of which needs a special DNA sequence called the
promoter and a set of DNA-binding proteins—
transcription factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fun ...
—to initiate the process (see regulation of transcription below).
RNA polymerase I
RNA polymerase 1 (also known as Pol I) is, in higher eukaryotes, the polymerase that only transcribes ribosomal RNA (but not 5S rRNA, which is synthesized by RNA polymerase III), a type of RNA that accounts for over 50% of the total RNA synthesiz ...
is responsible for transcription of ribosomal RNA (rRNA) genes.
RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ...
(Pol II) transcribes all protein-coding genes but also some non-coding RNAs (''e.g.'', snRNAs,
snoRNAs or
long non-coding RNA
Long non-coding RNAs (long ncRNAs, lncRNA) are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as mic ...
s).
RNA polymerase III
In eukaryote cells, RNA polymerase III (also called Pol III) is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA, and other small RNAs.
The genes transcribed by RNA Pol III fall in the category of "housekeeping" genes whose ex ...
transcribes
5S rRNA, transfer RNA (tRNA) genes, and some small non-coding RNAs (''e.g.'',
7SK). Transcription ends when the polymerase encounters a sequence called the
terminator.
mRNA processing
While transcription of prokaryotic protein-coding genes creates
messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA) that is ready for translation into protein, transcription of eukaryotic genes leaves a
primary transcript
A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNA ...
of RNA (pre-RNA), which first has to undergo a series of modifications to become a mature RNA. Types and steps involved in the maturation processes vary between coding and non-coding preRNAs; ''i.e.'' even though preRNA molecules for both mRNA and
tRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
undergo splicing, the steps and machinery involved are different. The processing of non-coding RNA is described below (non-coding RNA maturation).
The processing of pre-mRNA include 5′ ''capping'', which is set of enzymatic reactions that add
7-methylguanosine (m
7G) to the 5′ end of pre-mRNA and thus protect the RNA from degradation by
exonucleases
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolysis, hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the Directionality (molecular biolog ...
. The m
7G cap is then bound by
cap binding complex heterodimer (CBP20/CBP80), which aids in mRNA export to cytoplasm and also protect the RNA from
decapping.
Another modification is 3′ ''cleavage and polyadenylation''. They occur if polyadenylation signal sequence (5′- AAUAAA-3′) is present in pre-mRNA, which is usually between protein-coding sequence and terminator. The pre-mRNA is first cleaved and then a series of ~200 adenines (A) are added to form poly(A) tail, which protects the RNA from degradation. The poly(A) tail is bound by multiple
poly(A)-binding proteins (PABPs) necessary for mRNA export and translation re-initiation. In the inverse process of deadenylation, poly(A) tails are shortened by the
CCR4-Not
Carbon Catabolite Repression 4—Negative On TATA-less, or CCR4-Not, is a multiprotein complex that functions in gene expression. The complex has multiple enzymatic activities as both a poly(A) 3′-5′ exonuclease and a ubiquitin ligase. T ...
3′-5′ exonuclease, which often leads to full transcript decay.
A very important modification of eukaryotic pre-mRNA is ''
RNA splicing
RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcription (biology), transcript is transformed into a mature messenger RNA (Messenger RNA, mRNA). It works by removing all the introns (non-cod ...
''. The majority of eukaryotic pre-mRNAs consist of alternating segments called
exons
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence i ...
and
introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of ...
. During the process of splicing, an RNA-protein catalytical complex known as
spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to sp ...
catalyzes two
transesterification
Transesterification is the process of exchanging the organic functional group R″ of an ester with the organic group R' of an alcohol. These reactions are often catalyzed by the addition of an acid or base catalyst. Strong acids catalyze the r ...
reactions, which remove an intron and release it in form of lariat structure, and then splice neighbouring exons together. In certain cases, some introns or exons can be either removed or retained in mature mRNA. This so-called
alternative splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene ma ...
creates series of different transcripts originating from a single gene. Because these transcripts can be potentially translated into different proteins, splicing extends the complexity of eukaryotic gene expression and the size of a species
proteome
A proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. P ...
.
Extensive RNA processing may be an
evolutionary advantage made possible by the nucleus of eukaryotes. In prokaryotes, transcription and translation happen together, whilst in eukaryotes, the
nuclear membrane
The nuclear envelope, also known as the nuclear membrane, is made up of two lipid bilayer polar membrane, membranes that in eukaryotic cells surround the Cell nucleus, nucleus, which encloses the genome, genetic material.
The nuclear envelope con ...
separates the two processes, giving time for RNA processing to occur.
Non-coding RNA maturation
In most organisms
non-coding genes (ncRNA) are transcribed as precursors that undergo further processing. In the case of ribosomal RNAs (rRNA), they are often transcribed as a pre-rRNA that contains one or more rRNAs. The pre-rRNA is cleaved and modified (
2′-''O''-methylation and
pseudouridine formation) at specific sites by approximately 150 different small nucleolus-restricted RNA species, called snoRNAs. SnoRNAs associate with proteins, forming snoRNPs. While snoRNA part basepair with the target RNA and thus position the modification at a precise site, the protein part performs the catalytical reaction. In eukaryotes, in particular a snoRNP called RNase, MRP cleaves the
45S pre-rRNA into the
28S,
5.8S, and
18S rRNA
18S ribosomal RNA (abbreviated 18S rRNA) is a part of the ribosomal RNA in eukaryotes. It is a component of the Eukaryotic small ribosomal subunit (40S) and the cytosolic homologue of both the 12S rRNA in mitochondria and the 16S rRNA in plas ...
s. The rRNA and RNA processing factors form large aggregates called the
nucleolus
The nucleolus (; : nucleoli ) is the largest structure in the cell nucleus, nucleus of eukaryote, eukaryotic cell (biology), cells. It is best known as the site of ribosome biogenesis. The nucleolus also participates in the formation of signa ...
.
In the case of transfer RNA (tRNA), for example, the 5′ sequence is removed by
RNase P,
whereas the 3′ end is removed by the
tRNase Z enzyme
and the non-templated 3′ CCA tail is added by a
nucleotidyl transferase.
In the case of
micro RNA (miRNA), miRNAs are first transcribed as primary transcripts or pri-miRNA with a cap and poly-A tail and processed to short, 70-nucleotide stem-loop structures known as pre-miRNA in the cell nucleus by the enzymes
Drosha and
Pasha
Pasha (; ; ) was a high rank in the Ottoman Empire, Ottoman political and military system, typically granted to governors, generals, dignitary, dignitaries, and others. ''Pasha'' was also one of the highest titles in the 20th-century Kingdom of ...
. After being exported, it is then processed to mature miRNAs in the cytoplasm by interaction with the endonuclease
Dicer, which also initiates the formation of the
RNA-induced silencing complex (RISC), composed of the
Argonaute protein.
Even snRNAs and snoRNAs themselves undergo series of modification before they become part of functional RNP complex. This is done either in the nucleoplasm or in the specialized compartments called
Cajal bodies. Their bases are methylated or pseudouridinilated by a group of
small Cajal body-specific RNAs (scaRNAs), which are structurally similar to snoRNAs.
RNA export
In eukaryotes most mature RNA must be exported to the cytoplasm from the
nucleus. While some RNAs function in the nucleus, many RNAs are transported through the
nuclear pore
The nuclear pore complex (NPC), is a large protein complex giving rise to the nuclear pore. A great number of nuclear pores are studded throughout the nuclear envelope that surrounds the eukaryote cell nucleus. The pores enable the nuclear tran ...
s and into the
cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondri ...
.
Export of RNAs requires association with specific proteins known as exportins. Specific exportin molecules are responsible for the export of a given RNA type. mRNA transport also requires the correct association with
Exon Junction Complex (EJC), which ensures that correct processing of the mRNA is completed before export. In some cases RNAs are additionally transported to a specific part of the cytoplasm, such as a
synapse
In the nervous system, a synapse is a structure that allows a neuron (or nerve cell) to pass an electrical or chemical signal to another neuron or a target effector cell. Synapses can be classified as either chemical or electrical, depending o ...
; they are then towed by
motor protein
Motor proteins are a class of molecular motors that can move along the cytoskeleton of cells. They do this by converting chemical energy into mechanical work by the hydrolysis of ATP.
Cellular functions
Motor proteins are the driving force b ...
s that bind through linker proteins to specific sequences (called "zipcodes") on the RNA.
Translation
For some non-coding RNA, the mature RNA is the final gene product.
In the case of messenger RNA (mRNA) the RNA is an information carrier coding for the synthesis of one or more proteins. mRNA carrying a single protein sequence (common in eukaryotes) is
monocistronic whilst mRNA carrying multiple protein sequences (common in prokaryotes) is known as
polycistronic.

Every mRNA consists of three parts: a 5′ untranslated region (5′UTR), a protein-coding region or
open reading frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames ...
(ORF), and a 3′ untranslated region (3′UTR). The coding region carries information for protein synthesis encoded by the
genetic code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished ...
to form triplets. Each triplet of nucleotides of the
coding region is called a
codon
Genetic code is a set of rules used by living cells to translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links prote ...
and corresponds to a binding site complementary to an anticodon triplet in transfer RNA. Transfer RNAs with the same anticodon sequence always carry an identical type of
amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
. Amino acids are then chained together by the
ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
according to the order of triplets in the coding region. The ribosome helps transfer RNA to bind to messenger RNA and takes the amino acid from each transfer RNA and makes a structure-less protein out of it.
Each mRNA molecule is translated into many protein molecules, on average ~2800 in mammals.
In prokaryotes translation generally occurs at the point of transcription (co-transcriptionally), often using a messenger RNA that is still in the process of being created. In eukaryotes translation can occur in a variety of regions of the cell depending on where the protein being written is supposed to be. Major locations are the
cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
for soluble cytoplasmic proteins and the membrane of the
endoplasmic reticulum
The endoplasmic reticulum (ER) is a part of a transportation system of the eukaryote, eukaryotic cell, and has many other important functions such as protein folding. The word endoplasmic means "within the cytoplasm", and reticulum is Latin for ...
for proteins that are for export from the cell or insertion into a cell
membrane
A membrane is a selective barrier; it allows some things to pass through but stops others. Such things may be molecules, ions, or other small particles. Membranes can be generally classified into synthetic membranes and biological membranes. Bi ...
. Proteins that are supposed to be produced at the endoplasmic reticulum are recognised part-way through the translation process. This is governed by the
signal recognition particle
The signal recognition particle (SRP) is an abundant, cytosolic, universally conserved ribonucleoprotein (protein-RNA complex) that recognizes and targets specific proteins to the endoplasmic reticulum in eukaryotes and the plasma membrane ...
—a protein that binds to the ribosome and directs it to the endoplasmic reticulum when it finds a
signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16–30 amino acids long) present at the ...
on the growing (nascent) amino acid chain.
Folding
Each
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
exists as an unfolded
polypeptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ...
or random coil when translated from a sequence of
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
into a linear chain of
amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s. This polypeptide lacks any developed three-dimensional structure (the left hand side of the neighboring figure). The polypeptide then folds into its characteristic and functional
three-dimensional structure from a
random coil
In polymer chemistry, a random coil is a conformation of polymers where the monomer subunits are oriented randomly while still being bonded to adjacent units. It is not one specific shape, but a statistical distribution of shapes for all the cha ...
.
Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure) known as the
native state
In biochemistry, the native state of a protein or nucleic acid is its properly Protein folding, folded and/or assembled form, which is operative and functional. The native state of a biomolecule may possess all four levels of biomolecular structu ...
. The resulting three-dimensional structure is determined by the amino acid sequence (
Anfinsen's dogma).
The correct three-dimensional structure is essential to function, although some parts of functional proteins
may remain unfolded. Failure to fold into the intended shape usually produces inactive proteins with different properties including toxic
prion
A prion () is a Proteinopathy, misfolded protein that induces misfolding in normal variants of the same protein, leading to cellular death. Prions are responsible for prion diseases, known as transmissible spongiform encephalopathy (TSEs), w ...
s. Several
neurodegenerative and other
disease
A disease is a particular abnormal condition that adversely affects the structure or function (biology), function of all or part of an organism and is not immediately due to any external injury. Diseases are often known to be medical condi ...
s are believed to result from the accumulation of ''misfolded'' proteins.
Many
allergies
Allergies, also known as allergic diseases, are various conditions caused by hypersensitivity of the immune system to typically harmless substances in the environment. These diseases include Allergic rhinitis, hay fever, Food allergy, food al ...
are caused by the folding of the proteins, for the immune system does not produce antibodies for certain protein structures.
Enzymes called
chaperones assist the newly formed protein to attain (
fold into) the 3-dimensional structure it needs to function.
Similarly, RNA chaperones help RNAs attain their functional shapes.
Assisting protein folding is one of the main roles of the endoplasmic reticulum in eukaryotes.
Translocation
Secretory proteins of eukaryotes or prokaryotes must be translocated to enter the secretory pathway. Newly synthesized proteins are directed to the eukaryotic Sec61 or prokaryotic SecYEG translocation channel by
signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16–30 amino acids long) present at the ...
s. The efficiency of protein secretion in eukaryotes is very dependent on the
signal peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16–30 amino acids long) present at the ...
which has been used.
Protein transport
Many proteins are destined for other parts of the cell than the cytosol and a wide range of signalling sequences or
(signal peptides) are used to direct proteins to where they are supposed to be. In prokaryotes this is normally a simple process due to limited compartmentalisation of the cell. However, in eukaryotes there is a great variety of different targeting processes to ensure the protein arrives at the correct organelle.
Not all proteins remain within the cell and many are exported, for example,
digestive enzymes
Digestive enzymes take part in the chemical process of digestion, which follows the mechanical process of digestion. Food consists of macromolecules of proteins, carbohydrates, and fats that need to be broken down chemically by digestive enzymes ...
,
hormone
A hormone (from the Ancient Greek, Greek participle , "setting in motion") is a class of cell signaling, signaling molecules in multicellular organisms that are sent to distant organs or tissues by complex biological processes to regulate physio ...
s and
extracellular matrix
In biology, the extracellular matrix (ECM), also called intercellular matrix (ICM), is a network consisting of extracellular macromolecules and minerals, such as collagen, enzymes, glycoproteins and hydroxyapatite that provide structural and bio ...
proteins. In eukaryotes the export pathway is well developed and the main mechanism for the export of these proteins is translocation to the endoplasmic reticulum, followed by transport via the
Golgi apparatus
The Golgi apparatus (), also known as the Golgi complex, Golgi body, or simply the Golgi, is an organelle found in most eukaryotic Cell (biology), cells. Part of the endomembrane system in the cytoplasm, it protein targeting, packages proteins ...
.
Protein Degradation
Protein degradation is a major regulatory mechanism of gene expression and contributes substantially for shaping proteomes, especially of tissues and cells that do not grow very fast.
Protein degradation is a highly regulated processes, which results in significant and context dependent variation in degradation rates between proteins as well as for the same protein across cell types and tissue types. This variation can contribute about 40 % of the variance of protein levels across slowly growing tissues, with the remaining 60 % likely coming from protein synthesis, including transcription and translation as explained above.
Regulation of gene expression

Regulation of gene expression is the control of the amount and timing of appearance of the functional product of a gene. Control of expression is vital to allow a cell to produce the gene products it needs when it needs them; in turn, this gives cells the flexibility to adapt to a variable environment, external signals, damage to the cell, and other stimuli. More generally, gene regulation gives the cell control over all structure and function, and is the basis for
cellular differentiation
Cellular differentiation is the process in which a stem cell changes from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellula ...
,
morphogenesis
Morphogenesis (from the Greek ''morphê'' shape and ''genesis'' creation, literally "the generation of form") is the biological process that causes a cell, tissue or organism to develop its shape. It is one of three fundamental aspects of deve ...
and the versatility and adaptability of any organism.
Numerous terms are used to describe types of genes depending on how they are regulated; these include:
* A constitutive gene is a gene that is transcribed continually as opposed to a facultative gene, which is only transcribed when needed.
* A ''
housekeeping gene'' is a gene that is required to maintain basic cellular function and so is typically expressed in all cell types of an organism. Examples include
actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of ...
,
GAPDH and
ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 19 ...
. Some housekeeping genes are transcribed at a relatively constant rate and these genes can be used as a reference point in experiments to measure the expression rates of other genes.
* A facultative gene is a gene only transcribed when needed as opposed to a constitutive gene.
* An inducible gene is a gene whose expression is either responsive to environmental change or dependent on the position in the cell cycle.
Any step of gene expression may be modulated, from the DNA-RNA transcription step to
post-translational modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translation (biolog ...
of a protein. The stability of the final gene product, whether it is RNA or protein, also contributes to the expression level of the gene—an unstable product results in a low expression level. In general gene expression is regulated through changes
in the number and type of interactions between molecules
that collectively influence transcription of DNA
and translation of RNA.
Some simple examples of where gene expression is important are:
* Control of
insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the insulin (''INS)'' gene. It is the main Anabolism, anabolic hormone of the body. It regulates the metabol ...
expression so it gives a signal for
blood glucose regulation.
*
X chromosome inactivation in female
mammals
A mammal () is a vertebrate animal of the class Mammalia (). Mammals are characterised by the presence of milk-producing mammary glands for feeding their young, a broad neocortex region of the brain, fur or hair, and three middle e ...
to prevent an "overdose" of the genes it contains.
*
Cyclin
Cyclins are proteins that control the progression of a cell through the cell cycle by activating cyclin-dependent kinases (CDK).
Etymology
Cyclins were originally discovered by R. Timothy Hunt in 1982 while studying the cell cycle of sea urch ...
expression levels control progression through the eukaryotic
cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
.
Transcriptional regulation
 Regulation of transcription
Regulation of transcription can be broken down into three main routes of influence; genetic (direct interaction of a control factor with the gene), modulation interaction of a control factor with the transcription machinery and epigenetic (non-sequence changes in DNA structure that influence transcription).
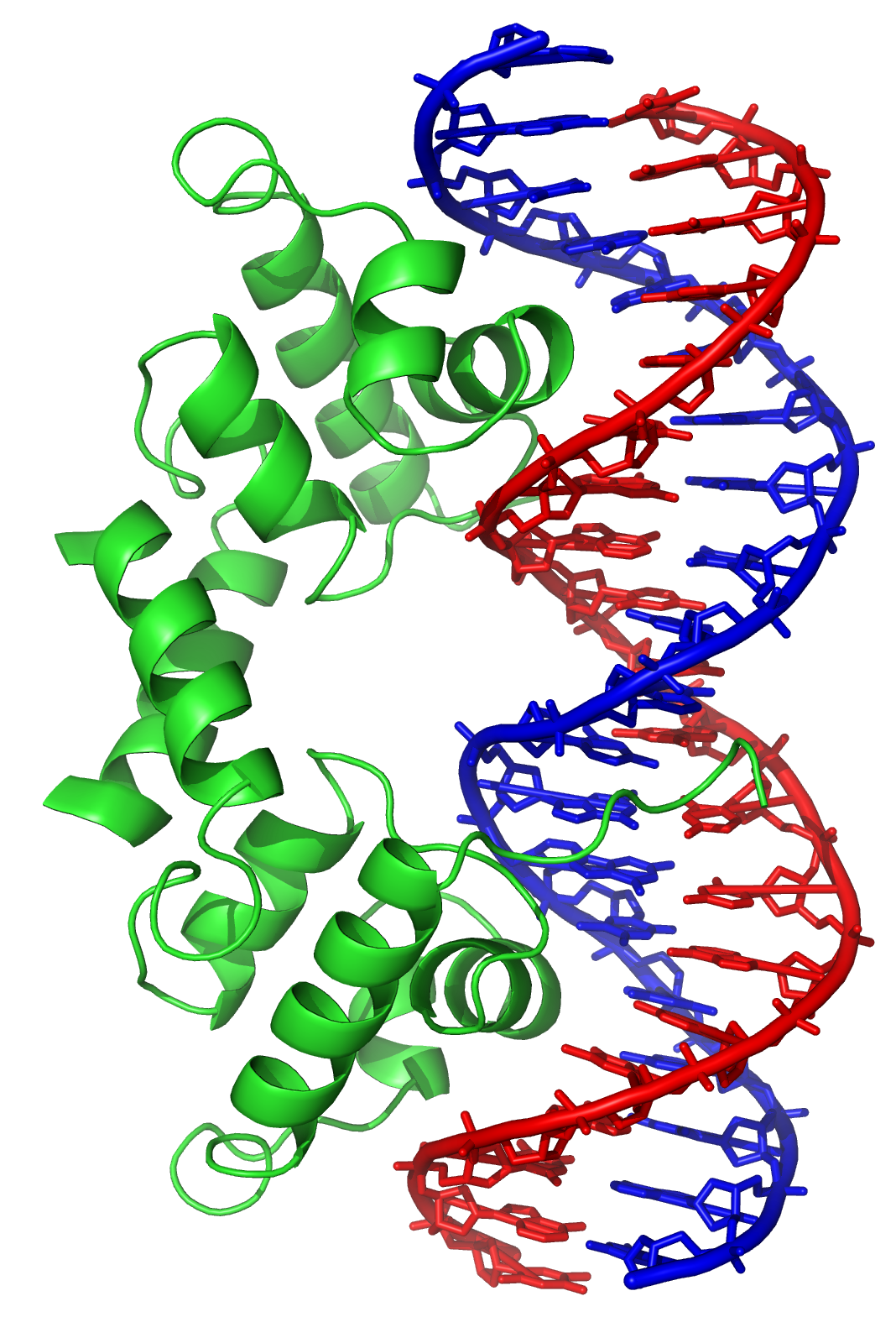
Direct interaction with DNA is the simplest and the most direct method by which a protein changes transcription levels. Genes often have several protein binding sites around the coding region with the specific function of regulating transcription. There are many classes of regulatory DNA binding sites known as
enhancers,
insulators and
silencers. The mechanisms for regulating transcription are varied, from blocking key binding sites on the DNA for
RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
to acting as an
activator and promoting transcription by assisting RNA polymerase binding.
The activity of transcription factors is further modulated by intracellular signals causing protein post-translational modification including
phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
,
acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
, or
glycosylation
Glycosylation is the reaction in which a carbohydrate (or ' glycan'), i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule (a glycosyl acceptor) in order to form a glycoconjugate. In biology (but not ...
. These changes influence a transcription factor's ability to bind, directly or indirectly, to promoter DNA, to recruit RNA polymerase, or to favor elongation of a newly synthesized RNA molecule.
The nuclear membrane in eukaryotes allows further regulation of transcription factors by the duration of their presence in the nucleus, which is regulated by reversible changes in their structure and by binding of other proteins.
Environmental stimuli or endocrine signals
may cause modification of regulatory proteins
eliciting cascades of intracellular signals,
which result in regulation of gene expression.
It has become apparent that there is a significant influence of non-DNA-sequence specific effects on transcription. These effects are referred to as
epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
and involve the higher order structure of DNA, non-sequence specific DNA binding proteins and chemical modification of DNA. In general epigenetic effects alter the accessibility of DNA to proteins and so modulate transcription.

In eukaryotes the structure of
chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
, controlled by the
histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
, regulates access to DNA with significant impacts on the expression of genes in
euchromatin
Euchromatin (also called "open chromatin") is a lightly packed form of chromatin (DNA, RNA, and protein) that is enriched in genes, and is often (but not always) under active transcription. Euchromatin stands in contrast to heterochromatin, which ...
and
heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
areas.
Enhancers, transcription factors, mediator complex and DNA loops in mammalian transcription

Gene expression in mammals is regulated by many
cis-regulatory element
''Cis''-regulatory elements (CREs) or ''cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morpho ...
s, including
core promoters and promoter-proximal elements that are located near the
transcription start sites of genes,
upstream on the DNA (towards the 5' region of the
sense strand). Other important cis-regulatory modules are localized in DNA regions that are distant from the transcription start sites. These include
enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
,
silencers,
insulators and tethering elements.
Enhancers and their associated
transcription factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fun ...
have a leading role in the regulation of gene expression.
Enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
are genome regions that regulate genes. Enhancers control cell-type-specific gene expression programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes.
Multiple enhancers, each often tens or hundred of thousands of nucleotides distant from their target genes, loop to their target gene promoters and coordinate with each other to control gene expression.
The illustration shows an enhancer looping around to come into proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of
CTCF
Transcriptional repressor CTCF also known as 11-zinc finger protein or CCCTC-binding factor is a transcription factor that in humans is encoded by the ''CTCF'' gene. CTCF is involved in many cellular processes, including transcriptional regulati ...
or
YY1). One member of the dimer is anchored to its binding motif on the enhancer and the other member is anchored to its binding motif on the promoter (represented by the red zigzags in the illustration).
Several cell function-specific transcription factors (among the about 1,600 transcription factors in a human cell)
generally bind to specific motifs on an enhancer.
A small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern transcription level of the target gene. Mediator (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (pol II) enzyme bound to the promoter.
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two eRNAs as illustrated in the figure.
An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of transcription factor bound to enhancer in the illustration).
An activated enhancer begins transcription of its RNA before activating transcription of messenger RNA from its target gene.
DNA methylation and demethylation in transcriptional regulation

DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
is a widespread mechanism for epigenetic influence on gene expression and is seen in
bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
and
eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
and has roles in heritable transcription silencing and transcription regulation. Methylation most often occurs on a cytosine (see Figure). Methylation of cytosine primarily occurs in dinucleotide sequences where a cytosine is followed by a guanine, a
CpG site. The number of
CpG sites in the human genome is about 28 million.
Depending on the type of cell, about 70% of the CpG sites have a methylated cytosine.
Methylation of cytosine in DNA has a major role in regulating gene expression. Methylation of CpGs in a promoter region of a gene usually represses gene transcription
while methylation of CpGs in the body of a gene increases expression.
play a central role in demethylation of methylated cytosines. Demethylation of CpGs in a gene promoter by
TET enzyme activity increases transcription of the gene.
Transcriptional regulation in learning and memory
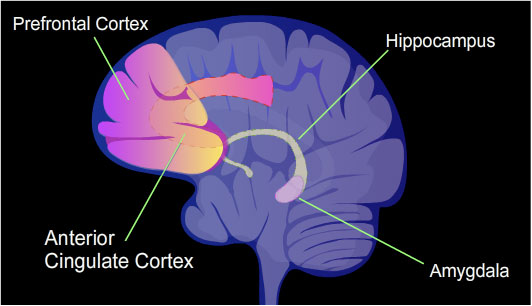
In a rat, contextual
fear conditioning
Pavlovian fear conditioning is a behavioral paradigm in which organisms learn to predict aversive events. It is a form of learning in which an aversive stimulus (e.g. an electrical shock) is associated with a particular neutral context (e.g., a r ...
(CFC) is a painful learning experience. Just one episode of CFC can result in a life-long fearful memory.
After an episode of CFC, cytosine methylation is altered in the promoter regions of about 9.17% of all genes in the
hippocampus
The hippocampus (: hippocampi; via Latin from Ancient Greek, Greek , 'seahorse'), also hippocampus proper, is a major component of the brain of humans and many other vertebrates. In the human brain the hippocampus, the dentate gyrus, and the ...
neuron DNA of a rat.
The
hippocampus
The hippocampus (: hippocampi; via Latin from Ancient Greek, Greek , 'seahorse'), also hippocampus proper, is a major component of the brain of humans and many other vertebrates. In the human brain the hippocampus, the dentate gyrus, and the ...
is where new memories are initially stored. After CFC about 500 genes have increased transcription (often due to demethylation of CpG sites in a promoter region) and about 1,000 genes have decreased transcription (often due to newly formed 5-methylcytosine at CpG sites in a promoter region). The pattern of induced and repressed genes within neurons appears to provide a molecular basis for forming the first transient memory of this training event in the hippocampus of the rat brain.
Some specific mechanisms guiding new DNA methylations and new
DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence ...
s in the
hippocampus
The hippocampus (: hippocampi; via Latin from Ancient Greek, Greek , 'seahorse'), also hippocampus proper, is a major component of the brain of humans and many other vertebrates. In the human brain the hippocampus, the dentate gyrus, and the ...
during memory establishment have been established (see
for summary). One mechanism includes guiding the short isoform of the
TET1 DNA demethylation
For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C) (see figure of 5mC and C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence ...
enzyme, TET1s, to about 600 locations on the genome. The guidance is performed by association of TET1s with
EGR1 protein, a transcription factor important in memory formation. Bringing TET1s to these locations initiates DNA demethylation at those sites, up-regulating associated genes. A second mechanism involves DNMT3A2, a splice-isoform of
DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl ...
DNMT3A, which adds methyl groups to cytosines in DNA. This isoform is induced by synaptic activity, and its location of action appears to be determined by
histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
post-translational modifications (a
histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
). The resulting new
messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
s are then transported by
messenger RNP particles (neuronal granules) to synapses of the neurons, where they can be translated into proteins affecting the activities of synapses.
In particular, the
brain-derived neurotrophic factor
Brain-derived neurotrophic factor (BDNF), or abrineurin, is a protein found in the and the periphery. that, in humans, is encoded by the ''BDNF'' gene. BDNF is a member of the neurotrophin family of growth factors, which are related to the can ...
gene (''BDNF'') is known as a "learning gene".
After CFC there was upregulation of ''BDNF'' gene expression, related to decreased CpG methylation of certain internal promoters of the gene, and this was correlated with learning.
Transcriptional regulation in cancer
The majority of gene
promoters contain a
CpG island with numerous
CpG sites.
When many of a gene's promoter CpG sites are
methylated
Methylation, in the chemical sciences, is the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These term ...
the gene becomes silenced.
Colorectal cancers typically have 3 to 6
driver mutations and 33 to 66
hitchhiker or passenger mutations.
However, transcriptional silencing may be of more importance than mutation in causing progression to cancer. For example, in colorectal cancers about 600 to 800 genes are transcriptionally silenced by CpG island methylation (see
regulation of transcription in cancer Generally, in progression to cancer, hundreds of genes are silenced or activated. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation (see DNA met ...
). Transcriptional repression in cancer can also occur by other
epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
mechanisms, such as altered expression of
microRNAs.
In breast cancer, transcriptional repression of
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a ...
may occur more frequently by over-transcribed microRNA-182 than by hypermethylation of the BRCA1 promoter (see
Low expression of BRCA1 in breast and ovarian cancers).
Post-transcriptional regulation
In eukaryotes, where export of RNA is required before translation is possible, nuclear export is thought to provide additional control over gene expression. All transport in and out of the nucleus is via the
nuclear pore
The nuclear pore complex (NPC), is a large protein complex giving rise to the nuclear pore. A great number of nuclear pores are studded throughout the nuclear envelope that surrounds the eukaryote cell nucleus. The pores enable the nuclear tran ...
and transport is controlled by a wide range of
importin
Importin is a type of karyopherin that transports protein molecules from the Eukaryotic Cell, cell's cytoplasm to the cell nucleus, nucleus. It does so by binding to specific recognition sequences, called nuclear localization sequences (NLS).
I ...
and
exportin proteins.
Expression of a gene coding for a protein is only possible if the messenger RNA carrying the code survives long enough to be translated.
In a typical cell, an RNA molecule is only stable if specifically protected from degradation. RNA degradation has particular importance in regulation of expression in eukaryotic cells where mRNA has to travel significant distances before being translated. In eukaryotes, RNA is stabilised by certain post-transcriptional modifications, particularly the
5′ cap and
poly-adenylated tail.
Intentional degradation of mRNA is used not just as a defence mechanism from foreign RNA (normally from viruses) but also as a route of mRNA ''destabilisation''. If an mRNA molecule has a complementary sequence to a
small interfering RNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA, double-stranded non-coding RNA, non-coding RNA, RNA molecules, typically 20–24 base pairs in length, similar to microR ...
then it is targeted for destruction via the
RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
pathway.
Three prime untranslated regions and microRNAs
Three prime untranslated regions (3′UTRs) of
messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
s (mRNAs) often contain regulatory sequences that post-transcriptionally influence gene expression. Such 3′-UTRs often contain both binding sites for
microRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcr ...
s (miRNAs) as well as for regulatory proteins. By binding to specific sites within the 3′-UTR, miRNAs can decrease gene expression of various mRNAs by either inhibiting translation or directly causing degradation of the transcript. The 3′-UTR also may have silencer regions that bind repressor proteins that inhibit the expression of a mRNA.
The 3′-UTR often contains
microRNA response elements (MREs). MREs are sequences to which miRNAs bind. These are prevalent motifs within 3′-UTRs. Among all regulatory motifs within the 3′-UTRs (e.g. including silencer regions), MREs make up about half of the motifs.
As of 2014, the
miRBase web site, an archive of
miRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcri ...
sequences
In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''terms''). The number of elements (possibly infinite) is call ...
and annotations, listed 28,645 entries in 233 biologic species. Of these, 1,881 miRNAs were in annotated human miRNA loci. miRNAs were predicted to have an average of about four hundred target
mRNAs (affecting expression of several hundred genes).
Friedman et al.
estimate that >45,000 miRNA target sites within human mRNA 3′UTRs are conserved above background levels, and >60% of human protein-coding genes have been under selective pressure to maintain pairing to miRNAs.
Direct experiments show that a single miRNA can reduce the stability of hundreds of unique mRNAs.
Other experiments show that a single miRNA may repress the production of hundreds of proteins, but that this repression often is relatively mild (less than 2-fold).
The effects of miRNA dysregulation of gene expression seem to be important in cancer.
For instance, in gastrointestinal cancers, nine miRNAs have been identified as
epigenetically altered and effective in down regulating DNA repair enzymes.
The effects of miRNA dysregulation of gene expression also seem to be important in neuropsychiatric disorders, such as schizophrenia, bipolar disorder, major depression, Parkinson's disease, Alzheimer's disease and autism spectrum disorders.
Translational regulation

Direct regulation of translation is less prevalent than control of transcription or mRNA stability but is occasionally used. Inhibition of protein translation is a major target for
toxin
A toxin is a naturally occurring poison produced by metabolic activities of living cells or organisms. They occur especially as proteins, often conjugated. The term was first used by organic chemist Ludwig Brieger (1849–1919), derived ...
s and
antibiotic
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting pathogenic bacteria, bacterial infections, and antibiotic medications are widely used in the therapy ...
s, so they can kill a cell by overriding its normal gene expression control.
Protein synthesis inhibitor
A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins.
While a broad interpretation of this definition could be used t ...
s include the antibiotic
neomycin
Neomycin, also known as framycetin, is an aminoglycoside antibiotic that displays bactericidal activity against Gram-negative aerobic bacilli and some anaerobic bacilli where resistance has not yet arisen. It is generally not effective against ...
and the toxin
ricin
Ricin ( ) is a lectin (a carbohydrate-binding protein) and a highly potent toxin produced in the seeds of the castor oil plant, ''Ricinus communis''. The median lethal dose (LD50) of ricin for mice is around 22 micrograms per kilogram of body ...
.
Post-translational modifications
Post-translational modifications (PTMs) are
covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
modifications to proteins. Like RNA splicing, they help to significantly diversify the proteome. These modifications are usually catalyzed by enzymes. Additionally, processes like covalent additions to amino acid side chain residues can often be reversed by other enzymes. However, some, like the
proteolytic cleavage of the protein backbone, are irreversible.
PTMs play many important roles in the cell.
For example, phosphorylation is primarily involved in activating and deactivating proteins and in signaling pathways.
PTMs are involved in transcriptional regulation: an important function of acetylation and methylation is histone tail modification, which alters how accessible DNA is for transcription.
They can also be seen in the immune system, where glycosylation plays a key role.
One type of PTM can initiate another type of PTM, as can be seen in how
ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 19 ...
tags proteins for degradation through proteolysis.
Proteolysis, other than being involved in breaking down proteins, is also important in activating and deactivating them, and in regulating biological processes such as DNA transcription and cell death.
Measurement

Measuring gene expression is an important part of many
life science
Life, also known as biota, refers to matter that has biological processes, such as signaling and self-sustaining processes. It is defined descriptively by the capacity for homeostasis, organisation, metabolism, growth, adaptation, respon ...
s, as the ability to quantify the level at which a particular gene is expressed within a cell, tissue or organism can provide a lot of valuable information. For example, measuring gene expression can:
* Identify viral infection of a cell (
viral protein
The term viral protein refers to both the products of the genome of a virus and any host proteins incorporated into the viral particle. Viral proteins are grouped according to their functions, and groups of viral proteins include structural protei ...
expression).
* Determine an individual's susceptibility to
cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
(
oncogene
An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels. expression).
* Find if a bacterium is resistant to
penicillin
Penicillins (P, PCN or PEN) are a group of beta-lactam antibiotic, β-lactam antibiotics originally obtained from ''Penicillium'' Mold (fungus), moulds, principally ''Penicillium chrysogenum, P. chrysogenum'' and ''Penicillium rubens, P. ru ...
(
beta-lactamase
Beta-lactamases (β-lactamases) are enzymes () produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems ( ertapenem), although carbapene ...
expression).
*
Gene expression profiling
In the field of molecular biology, gene expression profiling is the measurement of the activity (the gene expression, expression) of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, dis ...
evaluates a panel of genes to help understand the fundamental mechanism of a cell. This is increasingly used in cancer therapy to
target specific chemotherapy. (''See''
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
and
DNA_microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or t ...
for details.)
Similarly, the analysis of the location of protein expression is a powerful tool, and this can be done on an organismal or cellular scale. Investigation of localization is particularly important for the study of
development
Development or developing may refer to:
Arts
*Development (music), the process by which thematic material is reshaped
* Photographic development
*Filmmaking, development phase, including finance and budgeting
* Development hell, when a proje ...
in multicellular organisms and as an indicator of protein function in single cells. Ideally, measurement of expression is done by detecting the final gene product (for many genes, this is the protein); however, it is often easier to detect one of the precursors, typically
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
and to infer gene-expression levels from these measurements.
mRNA quantification
Levels of mRNA can be quantitatively measured by
northern blotting, which provides size and sequence information about the mRNA molecules. A sample of RNA is separated on an
agarose gel and hybridized to a radioactively labeled RNA probe that is complementary to the target sequence. The radiolabeled RNA is then detected by an
autoradiograph
An autoradiograph is an image on an X-ray film or nuclear emulsion produced by the pattern of decay emissions (e.g., beta particles or gamma rays) from a distribution of a radioactive substance. Alternatively, the autoradiograph is also availab ...
. Because the use of radioactive reagents makes the procedure time-consuming and potentially dangerous, alternative labeling and detection methods, such as digoxigenin and biotin chemistries, have been developed. Perceived disadvantages of Northern blotting are that large quantities of RNA are required and that quantification may not be completely accurate, as it involves measuring band strength in an image of a gel. On the other hand, the additional mRNA size information from the Northern blot allows the discrimination of alternately spliced transcripts.
Another approach for measuring mRNA abundance is RT-qPCR. In this technique,
reverse transcription
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B virus, hepatitis B to replicate their genomes, by retrot ...
is followed by
quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR when used quantitatively) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule duri ...
. Reverse transcription first generates a DNA template from the mRNA; this single-stranded template is called
cDNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engin ...
. The cDNA template is then amplified in the quantitative step, during which the
fluorescence
Fluorescence is one of two kinds of photoluminescence, the emission of light by a substance that has absorbed light or other electromagnetic radiation. When exposed to ultraviolet radiation, many substances will glow (fluoresce) with colore ...
emitted by labeled
hybridization probe
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA, usually 15–10000 nucleotides long, which can be radioactively or fluorescently labeled. HPs can be used to detect the presence of nucleotide sequences in analyzed ...
s or
intercalating dyes changes as the
DNA amplification process progresses. With a carefully constructed standard curve, qPCR can produce an absolute measurement of the number of copies of original mRNA, typically in units of copies per nanolitre of homogenized tissue or copies per cell. qPCR is very sensitive (detection of a single mRNA molecule is theoretically possible), but can be expensive depending on the type of reporter used; fluorescently labeled oligonucleotide probes are more expensive than non-specific intercalating fluorescent dyes.
For
expression profiling, or high-throughput analysis of many genes within a sample,
quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR when used quantitatively) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule duri ...
may be performed for hundreds of genes simultaneously in the case of low-density arrays. A second approach is the
hybridization microarray. A single array or "chip" may contain probes to determine transcript levels for every known gene in the genome of one or more organisms. Alternatively, "tag based" technologies like
Serial analysis of gene expression (SAGE) and
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
, which can provide a relative measure of the cellular
concentration
In chemistry, concentration is the abundance of a constituent divided by the total volume of a mixture. Several types of mathematical description can be distinguished: '' mass concentration'', '' molar concentration'', '' number concentration'', ...
of different mRNAs, can be used. An advantage of tag-based methods is the "open architecture", allowing for the exact measurement of any transcript, with a known or unknown sequence. Next-generation sequencing (NGS) such as
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
is another approach, producing vast quantities of sequence data that can be matched to a reference genome. Although NGS is comparatively time-consuming, expensive, and resource-intensive, it can identify
single-nucleotide polymorphisms
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in ...
, splice-variants, and novel genes, and can also be used to profile expression in organisms for which little or no sequence information is available.
RNA profiles in Wikipedia
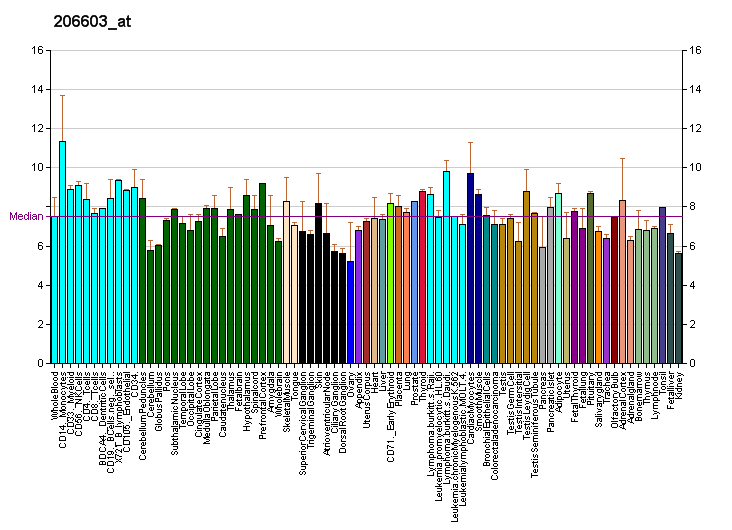
Profiles like these are found for almost all proteins listed in Wikipedia. They are generated by organizations such as the
Genomics Institute of the Novartis Research Foundation and the
European Bioinformatics Institute. Additional information can be found by searching their databases (for an example of the GLUT4 transporter pictured here, see citation). These profiles indicate the level of DNA expression (and hence RNA produced) of a certain protein in a certain tissue, and are color-coded accordingly in the images located in the Protein Box on the right side of each Wikipedia page.
Protein quantification
For genes encoding proteins, the expression level can be directly assessed by a number of methods with some clear analogies to the techniques for mRNA quantification.
One of the most commonly used methods is to perform a
Western blot
The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detect ...
against the protein of interest. This gives information on the size of the protein in addition to its identity. A sample (often cellular
lysate) is separated on a
polyacrylamide gel, transferred to a membrane and then probed with an
antibody
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as pathogenic bacteria, bacteria and viruses, includin ...
to the protein of interest. The antibody can either be conjugated to a
fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with se ...
or to
horseradish peroxidase
The enzyme horseradish peroxidase (HRP), found in the roots of horseradish, is used extensively in biochemistry applications. It is a metalloenzyme with many isoforms, of which the most studied type is C. It catalyzes the oxidation of various or ...
for imaging and/or quantification. The gel-based nature of this assay makes quantification less accurate, but it has the advantage of being able to identify later modifications to the protein, for example proteolysis or ubiquitination, from changes in size.
mRNA-protein correlation
While transcription directly reflects gene expression, the copy number of mRNA molecules does not directly correlate with the number of protein molecules translated from mRNA. Quantification of both protein and mRNA permits a correlation of the two levels. Regulation on each step of gene expression can impact the correlation, as shown for regulation of translation
or protein stability.
Post-translational factors, such as protein transport in highly polar cells,
can influence the measured mRNA-protein correlation as well.
Localization
Analysis of expression is not limited to quantification; localization can also be determined. mRNA can be detected with a suitably labelled complementary mRNA strand and protein can be detected via labelled antibodies. The probed sample is then observed by microscopy to identify where the mRNA or protein is.

By replacing the gene with a new version fused to a
green fluorescent protein
The green fluorescent protein (GFP) is a protein that exhibits green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea victo ...
marker or similar, expression may be directly quantified in live cells. This is done by imaging using a
fluorescence microscope
A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. A fluorescence micro ...
. It is very difficult to clone a GFP-fused protein into its native location in the genome without affecting expression levels, so this method often cannot be used to measure endogenous gene expression. It is, however, widely used to measure the expression of a gene artificially introduced into the cell, for example via an
expression vector
An expression vector, otherwise known as an expression construct, is usually a plasmid or virus designed for gene expression in cells. The vector (molecular biology), vector is used to introduce a specific gene into a target cell, and can command ...
. By fusing a target protein to a fluorescent reporter, the protein's behavior, including its cellular localization and expression level, can be significantly changed.
The
enzyme-linked immunosorbent assay works by using antibodies immobilised on a
microtiter plate to capture proteins of interest from samples added to the well. Using a detection antibody conjugated to an enzyme or fluorophore the quantity of bound protein can be accurately measured by
fluorometric or
colourimetric detection. The detection process is very similar to that of a Western blot, but by avoiding the gel steps more accurate quantification can be achieved.
Expression system
An expression system is a system specifically designed for the production of a gene product of choice. This is normally a protein although may also be RNA, such as
tRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
or a
ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozy ...
. An expression system consists of a gene, normally encoded by
DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
, and the
molecular machinery required to
transcribe the DNA into
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
and
translate the mRNA into
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
using the reagents provided. In the broadest sense this includes every living cell but the term is more normally used to refer to expression as a laboratory tool. An expression system is therefore often artificial in some manner. Expression systems are, however, a fundamentally natural process. Viruses are an excellent example where they replicate by using the host cell as an expression system for the viral proteins and genome.
Inducible expression
Doxycycline
Doxycycline is a Broad-spectrum antibiotic, broad-spectrum antibiotic of the Tetracycline antibiotics, tetracycline class used in the treatment of infections caused by bacteria and certain parasites. It is used to treat pneumonia, bacterial p ...
is also used in "Tet-on" and "Tet-off"
tetracycline controlled transcriptional activation to regulate
transgene
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques, from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the ...
expression in organisms and
cell culture
Cell culture or tissue culture is the process by which cell (biology), cells are grown under controlled conditions, generally outside of their natural environment. After cells of interest have been Cell isolation, isolated from living tissue, ...
s.
In nature
In addition to these biological tools, certain naturally observed configurations of DNA (genes, promoters, enhancers, repressors) and the associated machinery itself are referred to as an expression system. This term is normally used in the case where a gene or set of genes is switched on under well defined conditions, for example, the simple repressor switch expression system in
Lambda phage
Lambda phage (coliphage λ, scientific name ''Lambdavirus lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of ...
and the
lac operator system in bacteria. Several natural expression systems are directly used or modified and used for artificial expression systems such as the
Tet-on and Tet-off expression system.
Gene networks
Genes have sometimes been regarded as nodes in a network, with inputs being proteins such as
transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s, and outputs being the level of gene expression. The node itself performs a function, and the operation of these functions have been interpreted as performing a kind of information processing within cells and determines cellular behavior.
Gene networks can also be constructed without formulating an explicit causal model. This is often the case when assembling networks from large expression data sets. Covariation and correlation of expression is computed across a large sample of cases and measurements (often
transcriptome
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The ...
or
proteome
A proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. P ...
data). The source of variation can be either experimental or natural (observational). There are several ways to construct gene expression networks, but one common approach is to compute a matrix of all pair-wise correlations of expression across conditions, time points, or individuals and convert the matrix (after thresholding at some cut-off value) into a graphical representation in which nodes represent genes, transcripts, or proteins and edges connecting these nodes represent the strength of association (se
GeneNetwork GeneNetwork 2.
Techniques and tools
The following experimental techniques are used to measure gene expression and are listed in roughly chronological order, starting with the older, more established technologies. They are divided into two groups based on their degree of
multiplexity.
* Low-to-mid-plex techniques:
**
Reporter gene
**
Northern blot
**
Western blot
The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detect ...
**
Fluorescent in situ hybridization
Fluorescence ''in situ'' hybridization (FISH) is a cytogenetics, molecular cytogenetic technique that uses hybridization probe, fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence Com ...
**
Reverse transcription PCR
* Higher-plex techniques:
**
SAGE
**
DNA microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or t ...
**
Tiling array
**
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
Gene expression databases
Gene expression omnibus(GEO) at
NCBI
The National Center for Biotechnology Information (NCBI) is part of the National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is loca ...
Expression Atlasat the
EBI
Bgee Bgee at the
SIB Swiss Institute of Bioinformatics
* Mous
Gene Expression Databaseat the
Jackson Laboratory
*
CollecTF: a database of experimentally validated transcription factor-binding sites in Bacteria.
* COLOMBOS: collection of bacterial expression compendia.
Many Microbe Microarrays Database microbial Affymetrix data
See also
References
External links
Plant Transcription Factor Database and Plant Transcriptional Regulation Data and Analysis Platform
{{DEFAULTSORT:Gene Expression
Molecular biology
 Regulation of gene expression is the control of the amount and timing of appearance of the functional product of a gene. Control of expression is vital to allow a cell to produce the gene products it needs when it needs them; in turn, this gives cells the flexibility to adapt to a variable environment, external signals, damage to the cell, and other stimuli. More generally, gene regulation gives the cell control over all structure and function, and is the basis for
Regulation of gene expression is the control of the amount and timing of appearance of the functional product of a gene. Control of expression is vital to allow a cell to produce the gene products it needs when it needs them; in turn, this gives cells the flexibility to adapt to a variable environment, external signals, damage to the cell, and other stimuli. More generally, gene regulation gives the cell control over all structure and function, and is the basis for  Direct interaction with DNA is the simplest and the most direct method by which a protein changes transcription levels. Genes often have several protein binding sites around the coding region with the specific function of regulating transcription. There are many classes of regulatory DNA binding sites known as enhancers, insulators and silencers. The mechanisms for regulating transcription are varied, from blocking key binding sites on the DNA for
Direct interaction with DNA is the simplest and the most direct method by which a protein changes transcription levels. Genes often have several protein binding sites around the coding region with the specific function of regulating transcription. There are many classes of regulatory DNA binding sites known as enhancers, insulators and silencers. The mechanisms for regulating transcription are varied, from blocking key binding sites on the DNA for  In eukaryotes the structure of
In eukaryotes the structure of  Gene expression in mammals is regulated by many
Gene expression in mammals is regulated by many  In a rat, contextual
In a rat, contextual  Measuring gene expression is an important part of many
Measuring gene expression is an important part of many  Profiles like these are found for almost all proteins listed in Wikipedia. They are generated by organizations such as the Genomics Institute of the Novartis Research Foundation and the European Bioinformatics Institute. Additional information can be found by searching their databases (for an example of the GLUT4 transporter pictured here, see citation). These profiles indicate the level of DNA expression (and hence RNA produced) of a certain protein in a certain tissue, and are color-coded accordingly in the images located in the Protein Box on the right side of each Wikipedia page.
Profiles like these are found for almost all proteins listed in Wikipedia. They are generated by organizations such as the Genomics Institute of the Novartis Research Foundation and the European Bioinformatics Institute. Additional information can be found by searching their databases (for an example of the GLUT4 transporter pictured here, see citation). These profiles indicate the level of DNA expression (and hence RNA produced) of a certain protein in a certain tissue, and are color-coded accordingly in the images located in the Protein Box on the right side of each Wikipedia page.
 Analysis of expression is not limited to quantification; localization can also be determined. mRNA can be detected with a suitably labelled complementary mRNA strand and protein can be detected via labelled antibodies. The probed sample is then observed by microscopy to identify where the mRNA or protein is.
Analysis of expression is not limited to quantification; localization can also be determined. mRNA can be detected with a suitably labelled complementary mRNA strand and protein can be detected via labelled antibodies. The probed sample is then observed by microscopy to identify where the mRNA or protein is.
 By replacing the gene with a new version fused to a
By replacing the gene with a new version fused to a