|
Peptidyl Transferase
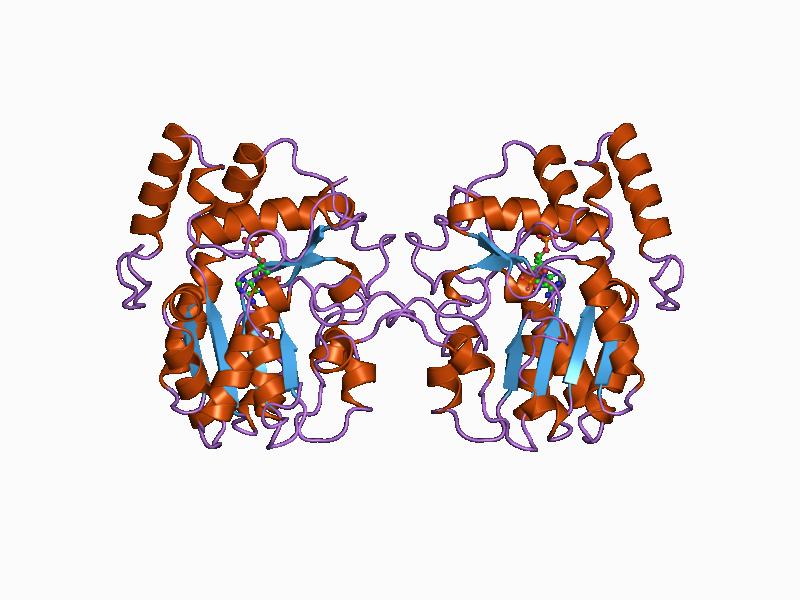
The peptidyl transferase center (, PTC) is an Aminoacyltransferases, aminoacyltransferase ribozyme (RNA enzyme) located in the large subunit of the ribosome. It forms peptide bonds between adjacent amino acids during the Translation (genetics), translation process of protein biosynthesis. It is also responsible for peptidyl-tRNA hydrolysis, allowing the release of the synthesized peptide chain at the end of translation. Peptidyl transferase activity is not mediated by any ribosomal proteins, but entirely by ribosomal RNA (rRNA). The catalytic activity of the PTC is a significant piece of evidence supporting the RNA World hypothesis. The PTC is a highly conserved region with a very slow rate of mutation. It is considered to be among the most ancient elements of the ribosome, probably predating the last universal common ancestor. The position of the PTC is analogous in all ribosomes (domain V in 23S numbering), being a part of the large subunit ribosomal RNA with the name only va ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aminoacyltransferases
Aminoacyltransferases () are acyltransferase enzymes which act upon an amino group. For instance, aminoacyl tRNA synthetases attach an aminoacid through esterification to the corresponding tRNA. The activation of amino acids it aminoacyl-tRNA synthetase requires hydrolysis of Adenosine triphosphate, ATP to Adenosine monophosphate, AMP plus Pyrophosphate, PPi. The aminoacyl-tRNA molecule has close relationships with elongation facts like EF-Tu. Peptidyl transferases are also a type of aminoacyltransferase that catalyze the formation of peptide bonds, as well as the hydrolytic step that leads to the release of newly synthesized proteins off the tRNA. External links * {{Portal bar, Biology, border=no EC 2.3.2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
P-site
The P-site (for peptidyl) is the second binding site for tRNA in the ribosome. The other two sites are the A-site (aminoacyl), which is the first binding site in the ribosome, and the E-site (exit), the third. During protein translation, the P-site holds the tRNA which is linked to the growing polypeptide chain. When a stop codon is reached, the peptidyl-tRNA bond of the tRNA located in the P-site is cleaved releasing the newly synthesized protein. During the translocation step of the elongation phase, the mRNA is advanced by one codon, coupled to movement of the tRNAs from the ribosomal A to P and P to E sites, catalyzed by elongation factor EF-G. Overview The ribosomal P-site plays a vital role in all phases of translation. Initiation involves recognition of the start codon (AUG) by initiator tRNA in the P-site, elongation involves passage of many elongator tRNAs through the P site, termination involves hydrolysis of the mature polypeptide from tRNA bound to the P-site, and ribos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transferase
In biochemistry, a transferase is any one of a class of enzymes that catalyse the transfer of specific functional groups (e.g. a methyl or glycosyl group) from one molecule (called the donor) to another (called the acceptor). They are involved in hundreds of different biochemical pathways throughout biology, and are integral to some of life's most important processes. Transferases are involved in myriad reactions in the cell. Three examples of these reactions are the activity of coenzyme A (CoA) transferase, which transfers thiol esters, the action of N-acetyltransferase, which is part of the pathway that metabolizes tryptophan, and the regulation of pyruvate dehydrogenase (PDH), which converts pyruvate to acetyl CoA. Transferases are also utilized during translation. In this case, an amino acid chain is the functional group transferred by a peptidyl transferase. The transfer involves the removal of the growing amino acid chain from the tRNA molecule in the A-site of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or Directed evolution, ''in vitro'' evolved ribozymes are the cleavage (or Ligation (molecular biology), ligation) of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3') can aminoacylate a GCCU-3' sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during Translation (biology), protein synthesis. They also participate ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Translocation (other)
{{disambig ...
Ribosomal translocation takes place in the elongation of a protein in: * * * Archaeal translation Archaeal translation is the process by which messenger RNA is translated into proteins in archaea. Not much is known on this subject, but on the protein level it seems to resemble eukaryotic translation. Most of the initiation, elongation, and ter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Macrolide
Macrolides are a class of mostly natural products with a large macrocyclic lactone ring to which one or more deoxy sugars, usually cladinose and desosamine, may be attached. Macrolides belong to the polyketide class of natural products. Some macrolides have antibiotic or antifungal activity and are used as pharmaceutical drugs. Rapamycin is also a macrolide and was originally developed as an antifungal, but has since been used as an immunosuppressant drug and is being investigated as a potential longevity therapeutic. Macrolides are a diverse group with many members of very different properties: * Macrolides with 14-, 15-, or 16-membered rings and two attached sugar molecules are antibiotics that bind to bacterial ribosomes, the key representative being erythromycin. The term "macrolide antibiotics" tend to refer to just this class. * Some macrolides with very large (20+ membered) rings are immunosuppresants, the prototypical one being rapamycin. * Some 23-membered ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
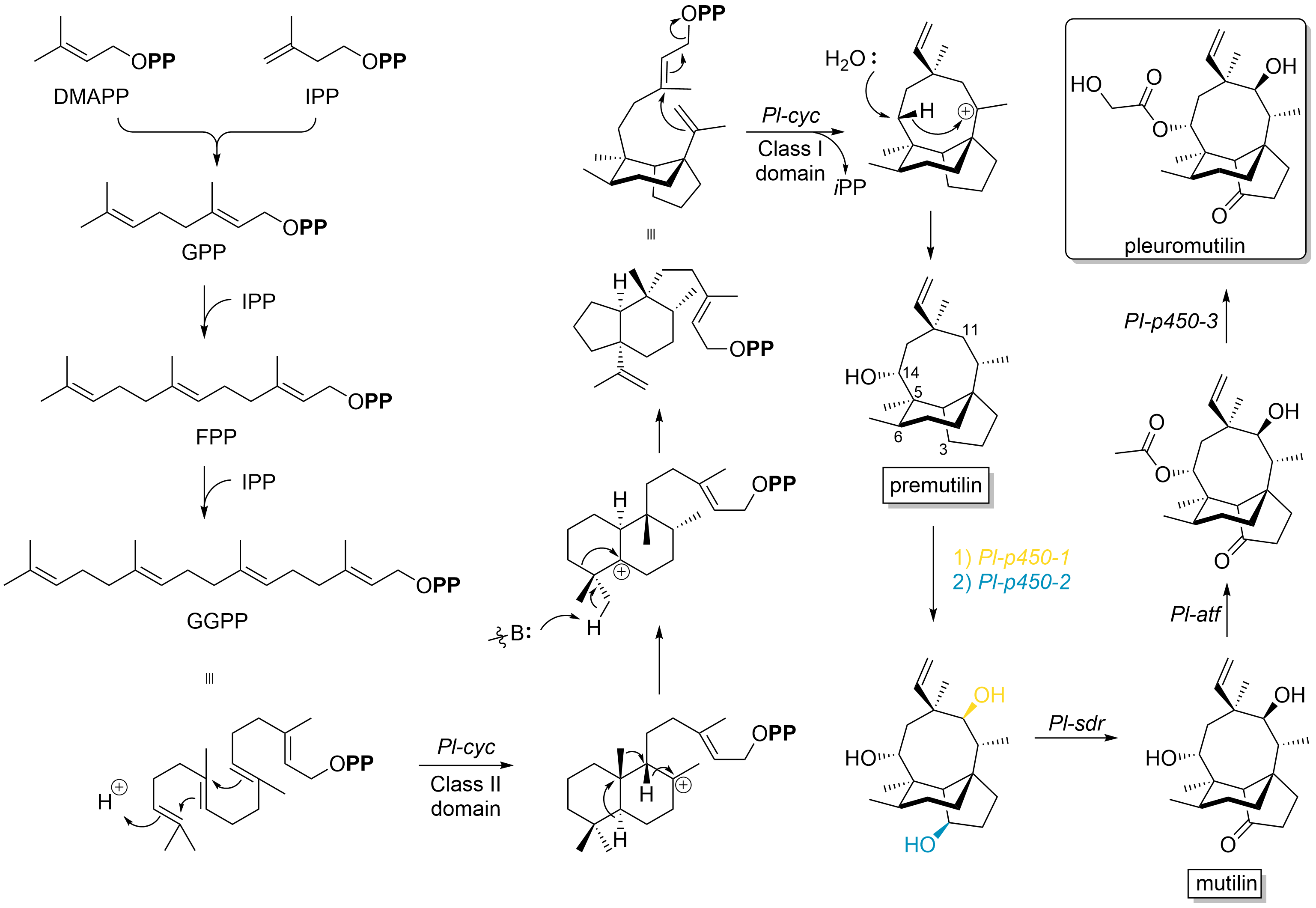
Pleuromutilin
Pleuromutilin and its derivatives are antibacterial drugs that inhibit protein synthesis in bacteria by binding to the peptidyl transferase component of the 50S subunit of ribosomes. This class of antibiotics includes the licensed drugs lefamulin (for systemic use in humans), retapamulin (approved for topical use in humans), valnemulin and tiamulin (approved for use in animals) and the investigational drug azamulin. History Pleuromutilin was discovered as an antibiotic in 1951. It is derived from the fungi '' Omphalina mutila'' (formerly ''Pleurotus mutilus'') and '' Clitopilus passeckerianus'' (formerly ''Pleurotus passeckerianus''), and has also been found in '' Drosophila subatrata'', '' Clitopilus scyphoides'', and some other '' Clitopilus'' species. Total synthesis The total synthesis of pleuromutilin has been reported. Biosynthesis Pleuromutilin belongs to the class of secondary metabolites known as terpenes, which are produced in fungi through the mevalonate pathw ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chloramphenicol
Chloramphenicol is an antibiotic useful for the treatment of a number of bacterial infections. This includes use as an eye ointment to treat conjunctivitis. By mouth or by intravenous, injection into a vein, it is used to treat meningitis, plague (disease), plague, cholera, and typhoid fever. Its use by mouth or by injection is only recommended when safer antibiotics cannot be used. Monitoring both blood levels of the medication and blood cell levels every two days is recommended during treatment. Common side effects include bone marrow suppression, nausea, and diarrhea. The bone marrow suppression may result in death. To reduce the risk of side effects treatment duration should be as short as possible. People with liver or kidney problems may need lower doses. In young infants, a condition known as gray baby syndrome may occur which results in a swollen stomach and Hypotension, low blood pressure. Its use near the end of pregnancy and during breastfeeding is typically not re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Synthesis Inhibitor
A protein synthesis inhibitor is a compound that stops or slows the growth or proliferation of cells by disrupting the processes that lead directly to the generation of new proteins. While a broad interpretation of this definition could be used to describe nearly any compound depending on concentration, in practice, it usually refers to compounds that act at the molecular level on translational machinery (either the ribosome itself or the translation factor), taking advantages of the major differences between prokaryotic and eukaryotic ribosome structures. Mechanism In general, protein synthesis inhibitors work at different stages of bacterial translation, bacterial mRNA translation into proteins, like initiation, elongation (including aminoacyl tRNA entry, Proofreading (biology), proofreading, peptidyl transfer, and bacterial translation#Elongation, bacterial translocation) and termination: Earlier stages * Rifamycin inhibits Bacterial transcription, bacterial DNA transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal Protein
A ribosomal protein (r-protein or rProtein) is any of the proteins that, in conjunction with rRNA, make up the ribosomal subunits involved in the cellular process of translation. ''E. coli'', other bacteria and Archaea have a 30S small subunit and a 50S large subunit, whereas humans and yeasts have a 40S small subunit and a 60S large subunit. Equivalent subunits are frequently numbered differently between bacteria, Archaea, yeasts and humans. A large part of the knowledge about these organic molecules has come from the study of '' E. coli'' ribosomes. All ribosomal proteins have been isolated and many specific antibodies have been produced. These, together with electronic microscopy and the use of certain reactives, have allowed for the determination of the topography of the proteins in the ribosome. More recently, a near-complete (near)atomic picture of the ribosomal proteins is emerging from the latest high-resolution cryo-EM data (including ). Conservation Ribosomal p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
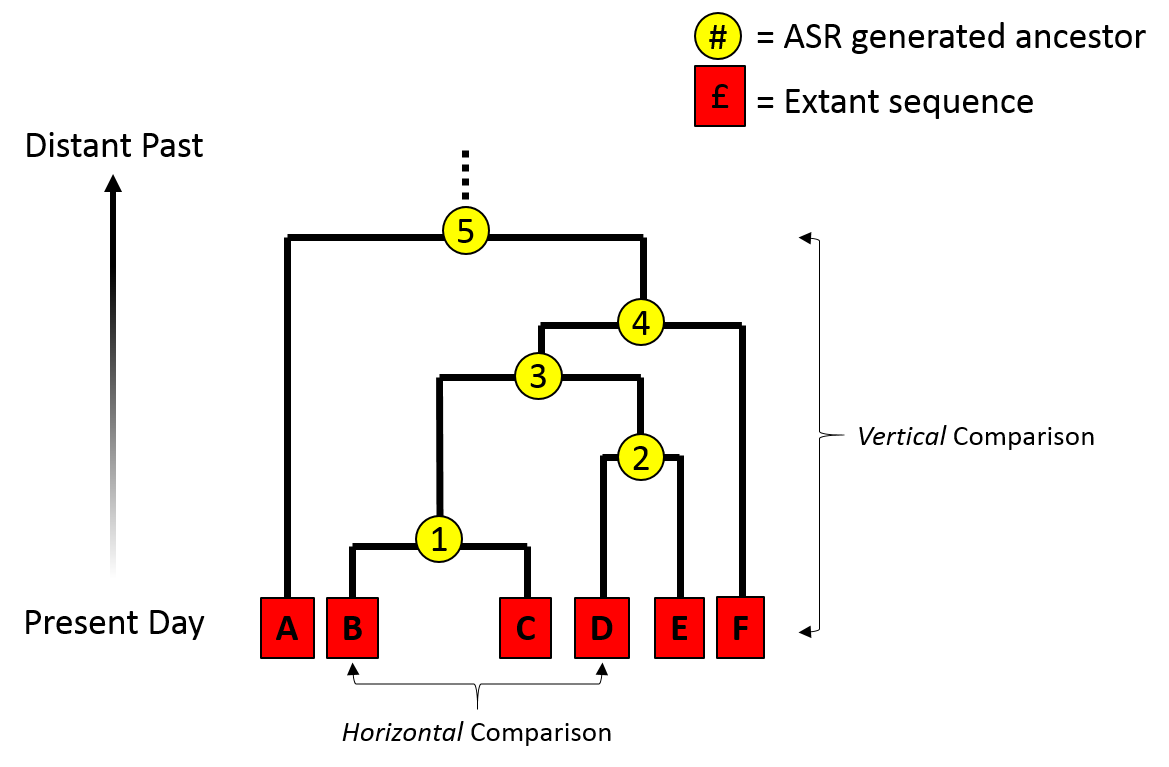
Ancestral Sequence Reconstruction
Ancestral sequence reconstruction (ASR) – also known as ancestral gene/sequence reconstruction/resurrection – is a technique used in the study of molecular evolution. The method uses related sequences to reconstruct an "ancestral" gene from a multiple sequence alignment. The method can be used to 'resurrect' ancestral proteins and was suggested in 1963 by Linus Pauling and Emile Zuckerkandl. In the case of enzymes, this approach has been called paleoenzymology (British: palaeoenzymology). Some early efforts were made in the 1980s and 1990s, led by the laboratory of Steven A. Benner, showing the potential of this technique. Thanks to the improvement of algorithms and of better sequencing and synthesis techniques, the method was developed further in the early 2000s to allow the resurrection of a greater variety of and much more ancient genes. Over the last decade, ancestral protein resurrection has developed as a strategy to reveal the mechanisms and dynamics of protein evolution ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |