ligand docking on:
[Wikipedia]
[Google]
[Amazon]
In the field of  The associations between biologically relevant molecules such as
The associations between biologically relevant molecules such as
 To perform a docking screen, the first requirement is a structure of the protein of interest. Usually the structure has been determined using a biophysical technique such as
*
To perform a docking screen, the first requirement is a structure of the protein of interest. Usually the structure has been determined using a biophysical technique such as
*
Docking@GRID
Project of Conformational Sampling and Docking on Grids : one aim is to deploy some intrinsic distributed docking algorithms on computational Grids, downloa
Docking@GRID open-source Linux version
Click2Drug.org
- Directory of computational drug design tools.
{{Webarchive, url=https://web.archive.org/web/20190202084025/http://www.chemcomp.com/MOE-Structure_Based_Design.htm#Ligand:ReceptorDocking , date=2019-02-02 with MOE (Molecular Operating Environment) Molecular modelling Computational chemistry Protein structure Medicinal chemistry Bioinformatics Drug discovery Articles containing video clips
molecular modeling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials scienc ...
, docking is a method which predicts the preferred orientation of one molecule to a second when a ligand
In coordination chemistry, a ligand is an ion or molecule with a functional group that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's el ...
and a target are bound to each other to form a stable complex
Complex commonly refers to:
* Complexity, the behaviour of a system whose components interact in multiple ways so possible interactions are difficult to describe
** Complex system, a system composed of many components which may interact with each ...
. Knowledge of the preferred orientation in turn may be used to predict the strength of association or binding affinity
In biochemistry and pharmacology, a ligand is a substance that forms a complex with a biomolecule to serve a biological purpose. The etymology stems from Latin ''ligare'', which means 'to bind'. In protein-ligand binding, the ligand is usuall ...
between two molecules using, for example, scoring functions SCORE may refer to:
*SCORE (software), a music scorewriter program
*SCORE (television), a weekend sports service of the defunct Financial News Network
*SCORE! Educational Centers
*SCORE International, an offroad racing organization
*Sarawak Corridor ...
.
 The associations between biologically relevant molecules such as
The associations between biologically relevant molecules such as proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, re ...
, peptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty am ...
s, nucleic acids
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic a ...
, carbohydrates
A carbohydrate () is a biomolecule composed of carbon (C), hydrogen (H), and oxygen (O) atoms. The typical hydrogen-to-oxygen atomic ratio is 2:1, analogous to that of water, and is represented by the empirical formula (where ''m'' and ''n'' ma ...
, and lipids
Lipids are a broad group of organic compounds which include fats, waxes, sterols, fat-soluble vitamins (such as vitamins Vitamin A, A, Vitamin D, D, Vitamin E, E and Vitamin K, K), monoglycerides, diglycerides, phospholipids, and others. The fu ...
play a central role in signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a biochemical cascade, series of molecular events. Proteins responsible for detecting stimuli are generally termed receptor (biology), rece ...
. Furthermore, the relative orientation of the two interacting partners may affect the type of signal produced (e.g., agonism
Agonism (from Greek 'struggle') is a political and social theory that emphasizes the potentially positive aspects of certain forms of conflict. It accepts a permanent place for such conflict in the political sphere, but seeks to show how indivi ...
vs antagonism). Therefore, docking is useful for predicting both the strength and type of signal produced.
Molecular docking is one of the most frequently used methods in structure-based drug design, due to its ability to predict the binding-conformation of small molecule
In molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight (≤ 1000 daltons) organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs are small molecules; ...
ligands to the appropriate target binding site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may includ ...
. Characterisation of the binding behaviour plays an important role in rational design of drugs as well as to elucidate fundamental biochemical processes. Hence, docking is useful to discover new ligand for a target by screening large virtual compound libraries and as a start for ligand optimization or investigation of mechanism of action.
Definition of problem
One can think of molecular docking as a problem of ''"lock-and-key"'', in which one wants to find the correct relative orientation of the ''"key"'' which will open up the ''"lock"'' (where on the surface of the lock is the key hole, which direction to turn the key after it is inserted, etc.). Here, the protein can be thought of as the "lock" and the ligand can be thought of as a "key". Molecular docking may be defined as an optimization problem, which would describe the "best-fit" orientation of a ligand that binds to a particular protein of interest. However, since both the ligand and the protein are flexible, a ''"hand-in-glove"'' analogy is more appropriate than ''"lock-and-key"''. During the course of the docking process, the ligand and the protein adjust their conformation to achieve an overall "best-fit" and this kind of conformational adjustment resulting in the overall binding is referred to as "induced-fit". Molecular docking research focuses on computationally simulating themolecular recognition
Supramolecular chemistry refers to the branch of chemistry concerning Chemical species, chemical systems composed of a integer, discrete number of molecules. The strength of the forces responsible for spatial organization of the system range from w ...
process. It aims to achieve an optimized conformation for both the protein and ligand and relative orientation between protein and ligand such that the free energy of the overall system is minimized.
Docking approaches
Two approaches are particularly popular within the molecular docking community. * One approach uses a matching technique that describes the protein and the ligand as complementary surfaces. * The second approach simulates the actual docking process in which the ligand-protein pairwise interaction energies are calculated. Both approaches have significant advantages as well as some limitations. These are outlined below.Shape complementarity
Geometric matching/shape complementarity methods describe the protein and ligand as a set of features that make them dockable. These features may include molecular surface/ complementary surface descriptors. In this case, the receptor's molecular surface is described in terms of its solvent-accessible surface area and the ligand's molecular surface is described in terms of its matching surface description. The complementarity between the two surfaces amounts to the shape matching description that may help finding the complementary pose of docking the target and the ligand molecules. Another approach is to describe the hydrophobic features of the protein using turns in the main-chain atoms. Yet another approach is to use a Fourier shape descriptor technique. Whereas the shape complementarity based approaches are typically fast and robust, they cannot usually model the movements or dynamic changes in the ligand/protein conformations accurately, although recent developments allow these methods to investigate ligand flexibility. Shape complementarity methods can quickly scan through several thousand ligands in a matter of seconds and actually figure out whether they can bind at the protein's active site, and are usually scalable to even protein-protein interactions. They are also much more amenable to pharmacophore based approaches, since they use geometric descriptions of the ligands to find optimal binding.Simulation
Simulating the docking process is much more complicated. In this approach, the protein and the ligand are separated by some physical distance, and the ligand finds its position into the protein's active site after a certain number of "moves" in its conformational space. The moves incorporate rigid body transformations such as translations and rotations, as well as internal changes to the ligand's structure including torsion angle rotations. Each of these moves in the conformation space of the ligand induces a total energetic cost of the system. Hence, the system's total energy is calculated after every move. The obvious advantage of docking simulation is that ligand flexibility is easily incorporated, whereas shape complementarity techniques must use ingenious methods to incorporate flexibility in ligands. Also, it more accurately models reality, whereas shape complementary techniques are more of an abstraction. Clearly, simulation is computationally expensive, having to explore a large energy landscape. Grid-based techniques, optimization methods, and increased computer speed have made docking simulation more realistic.Mechanics of docking
 To perform a docking screen, the first requirement is a structure of the protein of interest. Usually the structure has been determined using a biophysical technique such as
*
To perform a docking screen, the first requirement is a structure of the protein of interest. Usually the structure has been determined using a biophysical technique such as
* X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
,
* NMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique based on re-orientation of atomic nuclei with non-zero nuclear spins in an external magnetic f ...
or
* cryo-electron microscopy (cryo-EM),
but can also derive from homology modeling
Homology modeling, also known as comparative modeling of protein, refers to constructing an atomic-resolution model of the "''target''" protein from its amino acid sequence and an experimental three-dimensional structure of a related homologous pr ...
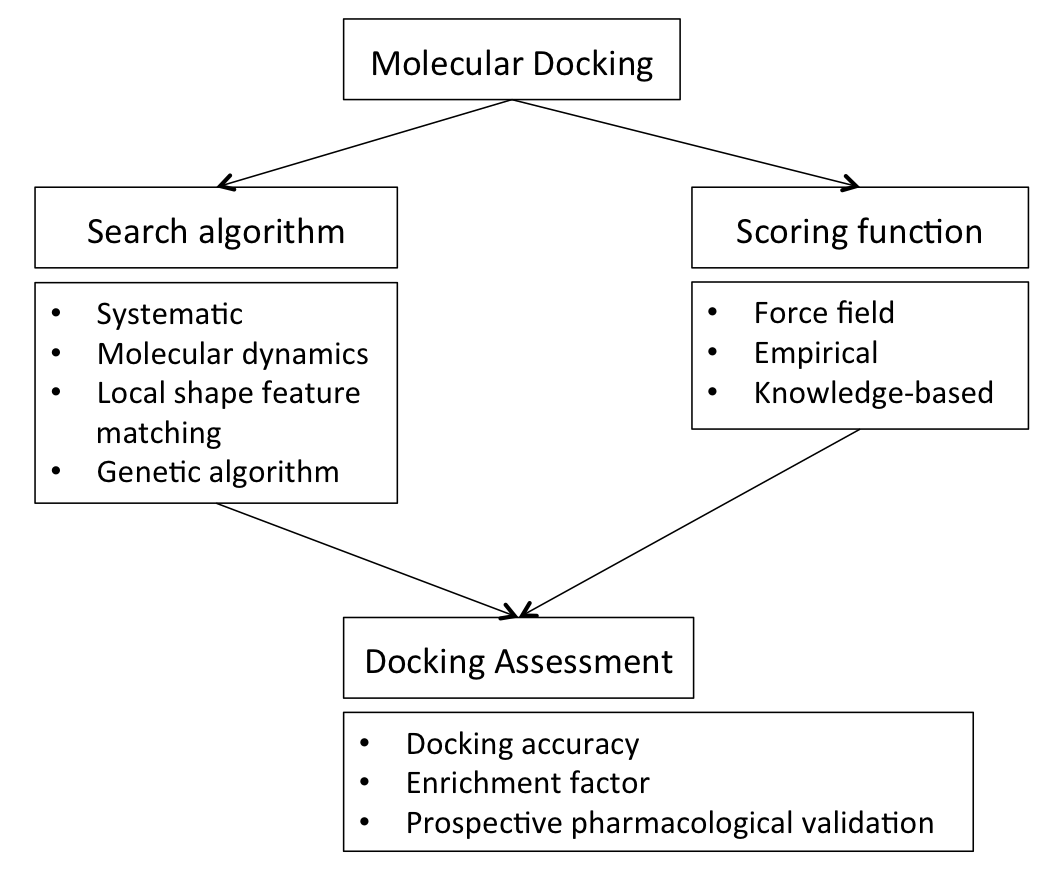
construction. This protein structure and a database of potential ligands serve as inputs to a docking program. The success of a docking program depends on two components: the search algorithm
In computer science, a search algorithm is an algorithm designed to solve a search problem. Search algorithms work to retrieve information stored within particular data structure, or calculated in the Feasible region, search space of a problem do ...
and the scoring function.
Search algorithm
The search space in theory consists of all possible orientations and conformations of the protein paired with the ligand. However, in practice with current computational resources, it is impossible to exhaustively explore the search space — this would involve enumerating all possible distortions of each molecule (molecules are dynamic and exist in an ensemble of conformational states) and all possiblerotational
Rotation or rotational/rotary motion is the circular movement of an object around a central line, known as an ''axis of rotation''. A plane figure can rotate in either a clockwise or counterclockwise sense around a perpendicular axis intersec ...
and translational orientations of the ligand relative to the protein at a given level of granularity
Granularity (also called graininess) is the degree to which a material or system is composed of distinguishable pieces, "granules" or "grains" (metaphorically).
It can either refer to the extent to which a larger entity is subdivided, or the ...
. Most docking programs in use account for the whole conformational space of the ligand (flexible ligand), and several attempt to model a flexible protein receptor. Each "snapshot" of the pair is referred to as a pose.
A variety of conformational search strategies have been applied to the ligand and to the receptor. These include:
* systematic or stochastic Stochastic (; ) is the property of being well-described by a random probability distribution. ''Stochasticity'' and ''randomness'' are technically distinct concepts: the former refers to a modeling approach, while the latter describes phenomena; i ...
torsional
In the field of solid mechanics, torsion is the twisting of an object due to an applied torque. Torsion could be defined as strain or angular deformation, and is measured by the angle a chosen section is rotated from its equilibrium position. Th ...
searches about rotatable bonds
* molecular dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the Motion (physics), physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamics ( ...
simulations
* genetic algorithm
In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to g ...
s to "evolve" new low energy conformations and where the score of each pose acts as the fitness function used to select individuals for the next iteration.
Ligand flexibility
Conformations of the ligand may be generated in the absence of the receptor and subsequently docked or conformations may be generated on-the-fly in the presence of the receptor binding cavity, or with full rotational flexibility of every dihedral angle using fragment based docking. Force field energy evaluation are most often used to select energetically reasonable conformations, but knowledge-based methods have also been used. Peptides are both highly flexible and relatively large-sized molecules, which makes modeling their flexibility a challenging task. A number of methods were developed to allow for efficient modeling of flexibility of peptides during protein-peptide docking.Receptor flexibility
Computational capacity has increased dramatically over the last decade making possible the use of more sophisticated and computationally intensive methods in computer-assisted drug design. However, dealing with receptor flexibility in docking methodologies is still a thorny issue. The main reason behind this difficulty is the large number of degrees of freedom that have to be considered in this kind of calculations. Neglecting it, however, in some of the cases may lead to poor docking results in terms of binding pose prediction. Multiple static structures experimentally determined for the same protein in different conformations are often used to emulate receptor flexibility. Alternatively rotamer libraries of amino acid side chains that surround the binding cavity may be searched to generate alternate but energetically reasonable protein conformations.Scoring function
Docking programs generate a large number of potential ligand poses, of which some can be immediately rejected due to clashes with the protein. The remainder are evaluated using some scoring function, which takes a pose as input and returns a number indicating the likelihood that the pose represents a favorable binding interaction and ranks one ligand relative to another. Most scoring functions are physics-basedmolecular mechanics
Molecular mechanics uses classical mechanics to model molecular systems. The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using Force field (chemi ...
force fields that estimate the energy of the pose within the binding site. The various contributions to binding can be written as an additive equation:
The components consist of solvent effects, conformational changes in the protein and ligand, free energy due to protein-ligand interactions, internal rotations, association energy of ligand and receptor to form a single complex and free energy due to changes in vibrational modes. A low (negative) energy indicates a stable system and thus a likely binding interaction.
Alternative approaches use modified scoring functions to include constraints based on known key protein-ligand interactions, or knowledge-based potentials derived from interactions observed in large databases of protein-ligand structures (e.g. the Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained a ...
).
There are a large number of structures from X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
for complexes between proteins and high affinity ligands, but comparatively fewer for low affinity ligands as the latter complexes tend to be less stable and therefore more difficult to crystallize. Scoring functions trained with this data can dock high affinity ligands correctly, but they will also give plausible docked conformations for ligands that do not bind. This gives a large number of false positive
A false positive is an error in binary classification in which a test result incorrectly indicates the presence of a condition (such as a disease when the disease is not present), while a false negative is the opposite error, where the test resu ...
hits, i.e., ligands predicted to bind to the protein that actually don't when placed together in a test tube.
One way to reduce the number of false positives is to recalculate the energy of the top scoring poses using (potentially) more accurate but computationally more intensive techniques such as Generalized Born or Poisson-Boltzmann methods.
Docking assessment
The interdependence between sampling and scoring function affects the docking capability in predicting plausible poses or binding affinities for novel compounds. Thus, an assessment of a docking protocol is generally required (when experimental data is available) to determine its predictive capability. Docking assessment can be performed using different strategies, such as: * docking accuracy (DA) calculation; * the correlation between a docking score and the experimental response or determination of the enrichment factor (EF); * the distance between an ion-binding moiety and the ion in the active site; * the presence of induce-fit models.Docking accuracy
Docking accuracy represents one measure to quantify the fitness of a docking program by rationalizing the ability to predict the right pose of a ligand with respect to that experimentally observed.Enrichment factor
Docking screens can also be evaluated by the enrichment of annotated ligands of known binders from among a large database of presumed non-binding, "decoy
A decoy (derived from the Dutch ''de'' ''kooi'', literally "the cage" or possibly ''eenden kooi'', " duck cage") is usually a person, device, or event which resembles what an individual or a group might be looking for, but it is only meant to ...
" molecules. In this way, the success of a docking screen is evaluated by its capacity to enrich the small number of known active compounds in the top ranks of a screen from among a much greater number of decoy molecules in the database. The area under the receiver operating characteristic (ROC) curve is widely used to evaluate its performance.
Prospective
Resulting hits from docking screens are subjected to pharmacological validation (e.g. IC50,affinity
Affinity may refer to:
Commerce, finance and law
* Affinity (law), kinship by marriage
* Affinity analysis, a market research and business management technique
* Affinity Credit Union, a Saskatchewan-based credit union
* Affinity Equity Pa ...
or potency measurements). Only prospective studies constitute conclusive proof of the suitability of a technique for a particular target. In the case of G protein-coupled receptors
G protein-coupled receptors (GPCRs), also known as seven-(pass)-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptors, and G protein-linked receptors (GPLR), form a large protein family, group of evoluti ...
(GPCRs), which are targets of more than 30% of marketed drugs, molecular docking led to the discovery of more than 500 GPCR ligands.
Benchmarking
The potential of docking programs to reproduce binding modes as determined byX-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
can be assessed by a range of docking benchmark sets.
For small molecules, several benchmark data sets for docking and virtual screening exist e.g. ''Astex Diverse Set'' consisting of high quality protein−ligand X-ray crystal structures, the ''Directory of Useful Decoys'' (DUD) for evaluation of virtual screening performance, or the LEADS-FRAG data set for fragments
An evaluation of docking programs for their potential to reproduce peptide binding modes can be assessed by ''Lessons for Efficiency Assessment of Docking and Scoring'' (LEADS-PEP).
Applications
A binding interaction between asmall molecule
In molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight (≤ 1000 daltons) organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs are small molecules; ...
ligand and an enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
protein may result in activation or inhibition
Inhibitor or inhibition may refer to:
Biology
* Enzyme inhibitor, a substance that binds to an enzyme and decreases the enzyme's activity
* Reuptake inhibitor, a substance that increases neurotransmission by blocking the reuptake of a neurotransm ...
of the enzyme. If the protein is a receptor, ligand binding may result in agonism
Agonism (from Greek 'struggle') is a political and social theory that emphasizes the potentially positive aspects of certain forms of conflict. It accepts a permanent place for such conflict in the political sphere, but seeks to show how indivi ...
or antagonism. Docking is most commonly used in the field of drug design
Drug design, often referred to as rational drug design or simply rational design, is the invention, inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic compound, organi ...
— most drugs are small organic molecules, and docking may be applied to:
* hit identification – docking combined with a scoring function can be used to quickly screen large databases of potential drugs in silico
In biology and other experimental sciences, an ''in silico'' experiment is one performed on a computer or via computer simulation software. The phrase is pseudo-Latin for 'in silicon' (correct ), referring to silicon in computer chips. It was c ...
to identify molecules that are likely to bind to protein target of interest (see virtual screening
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor (biochemistry), r ...
). Reverse pharmacology routinely uses docking for target identification.
* lead optimization – docking can be used to predict in where and in which relative orientation a ligand binds to a protein (also referred to as the binding mode or pose). This information may in turn be used to design more potent and selective analogs.
* bioremediation
Bioremediation broadly refers to any process wherein a biological system (typically bacteria, microalgae, fungi in mycoremediation, and plants in phytoremediation), living or dead, is employed for removing environmental pollutants from air, wate ...
– protein ligand docking can also be used to predict pollutants that can be degraded by enzymes.
See also
*Drug design
Drug design, often referred to as rational drug design or simply rational design, is the invention, inventive process of finding new medications based on the knowledge of a biological target. The drug is most commonly an organic compound, organi ...
* Katchalski-Katzir algorithm
* List of molecular graphics systems
* Macromolecular docking
* Molecular mechanics
Molecular mechanics uses classical mechanics to model molecular systems. The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using Force field (chemi ...
* Protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid ...
* Protein design
* Software for molecular mechanics modeling
* List of protein-ligand docking software
* Molecular design software Molecular design software is notable software for molecular modeling, that provides special support for developing molecular models ''de novo''.
In contrast to the usual molecular modeling programs, such as for molecular dynamics and quantum chemi ...
* Docking@Home
* Exscalate4Cov
* Ibercivis
* ZINC database
* Lead Finder
* Virtual screening
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor (biochemistry), r ...
* Scoring functions for docking
* Ultra-large-scale docking
References
External links
* *Docking@GRID
Project of Conformational Sampling and Docking on Grids : one aim is to deploy some intrinsic distributed docking algorithms on computational Grids, downloa
Docking@GRID open-source Linux version
Click2Drug.org
- Directory of computational drug design tools.
{{Webarchive, url=https://web.archive.org/web/20190202084025/http://www.chemcomp.com/MOE-Structure_Based_Design.htm#Ligand:ReceptorDocking , date=2019-02-02 with MOE (Molecular Operating Environment) Molecular modelling Computational chemistry Protein structure Medicinal chemistry Bioinformatics Drug discovery Articles containing video clips