|
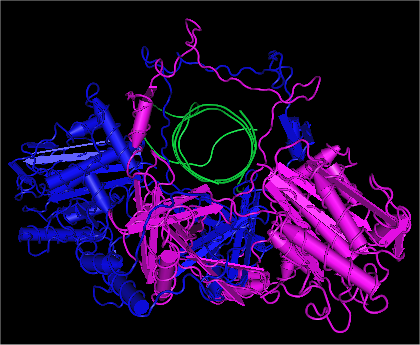
Ku70
Ku70 is a heterodimeric protein made up of Ku70 and Ku80, which together form Ku. In humans, is encoded by the ''XRCC6'' gene. Ku70 plays a critical role in the DNA repair, maintenance and many other cellular processes. Function Together, Ku70 and Ku80 make up the Ku heterodimer form a quasi-symmetric structure, which encircles the double-stranded DNA. The DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) of the DNA repair pathway. It is also required for V(D)J recombination, which utilizes the NHEJ pathway to promote antigen diversity in the mammalian immune system. Ku70 is key for sensing and responding to cytosolic DNA, which is essential for the indication of infection. Within the heterodimer, Ku70 specifically binds directly to broken ends of double-stranded DNA breaks, or DSBs. Then together, Ku70 and Ku80 will tightly form a ring-like structure around the DNA strand, preventing further degradation. These steps are essential for the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ku (protein)
Ku is a dimeric protein complex that binds to DNA double-strand break DNA end, ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. Ku is evolutionarily conserved from bacteria to humans. The ancestral bacterial Ku is a homodimer (two copies of the same protein bound to each other). Eukaryotic Ku is a heterodimer of two polypeptides, Ku70 (XRCC6) and Ku80 (XRCC5), so named because the molecular weight of the human Ku proteins is around 70 kDa and 80 kDa. The two Ku subunits form a basket-shaped structure that threads onto the DNA end. Once bound, Ku can slide down the DNA strand, allowing more Ku molecules to thread onto the end. In higher eukaryotes, Ku forms a complex with the DNA-PKcs, DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form the full DNA-dependent protein kinase, DNA-PK. Ku is thought to function as a molecular scaffold to which other proteins involved in NHEJ can bind, orienting the double-strand break for ligat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ku80
Ku80 is a protein that, in humans, is encoded by the ''XRCC5'' gene. Together, Ku70 and Ku80 make up the Ku heterodimer, which binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. It is also required for V(D)J recombination, which utilizes the NHEJ pathway to promote antigen diversity in the mammalian immune system. In addition to its role in NHEJ, Ku is required for telomere length maintenance and subtelomeric gene silencing. Ku was originally identified when patients with systemic lupus erythematosus were found to have high levels of autoantibodies to the protein. Nomenclature Ku80 has been referred to by several names including: * Lupus Ku autoantigen protein p80 * ATP-dependent DNA helicase 2 subunit 2 * X-ray repair complementing defective repair in Chinese hamster cells 5 * X-ray repair cross-complementing 5 (XRCC5) Epigenetic repression The protein expression level of Ku80 can be repressed by e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage Theory Of Aging
The DNA damage theory of aging proposes that aging is a consequence of unrepaired accumulation of DNA damage (naturally occurring), naturally occurring DNA damage. Damage in this context is a DNA alteration that has an abnormal structure. Although both mitochondrion, mitochondrial and Cell nucleus, nuclear DNA damage can contribute to aging, nuclear DNA is the main subject of this analysis. Nuclear DNA damage can contribute to aging either indirectly (by increasing apoptosis or Hayflick limit, cellular senescence) or directly (by increasing cell dysfunction). Several review articles have shown that deficient DNA repair, allowing greater accumulation of DNA damage, causes premature aging; and that increased DNA repair facilitates greater longevity, e.g. Mouse models of nucleotide-excision–repair syndromes reveal a striking correlation between the degree to which specific DNA repair pathways are compromised and the severity of accelerated aging, strongly suggesting a causal relation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair (HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallmarks of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TERF2
Telomeric repeat-binding factor 2 is a protein that is present at telomeres throughout the cell cycle. It is also known as TERF2, TRF2, and TRBF2, and is encoded in humans by the ''TERF2'' gene. It is a component of the shelterin nucleoprotein complex and a second negative regulator of telomere length, playing a key role in the protective activity of telomeres. It was first reported in 1997 in the lab of Titia de Lange, where a DNA sequence similar, but not identical, to TERF1 was discovered, with respect to the Myb-domain. De Lange isolated the new Myb-containing protein sequence and called it TERF2. Structure and domains TERF2 has a similar structure to that of TERF1. Both proteins carry a C-terminus Myb motif and large TERF1-related dimerization domains near their N-terminus. However, both proteins exist exclusively as homodimers and do not heterodimerize with each other, as proven by co-immunoprecipitation assay analysis. Also, TERF2 has a basic N-terminus, differing fr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CBX5 (gene)
Chromobox protein homolog 5 is a protein that in humans is encoded by the ''CBX5'' gene. It is a highly conserved, non-histone protein part of the heterochromatin family. The protein itself is more commonly called (in humans) HP1α. Heterochromatin protein-1 (HP1) has an N-terminal domain that acts on methylated lysines residues leading to epigenetic repression. The C-terminal of this protein has a chromo shadow-domain (CSD) that is responsible for homodimerizing, as well as interacting with a variety of chromatin-associated, non-histone proteins. Structure HP1α is 191 amino acids in length containing 6 exons. As mentioned above, this protein contains two domains, an N-terminal chromodomain (CD) and a C- terminal chromoshadow domain (CSD). The CD binds with histone 3 through a methylated lysine residue at position 9 (H3K9) while the C-terminal CSD homodimerizes and interacts with a variety of other chromatin-associated, non-histone related proteins. Connecting these two domai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MRE11A
Double-strand break repair protein MRE11 (Meiotic recombination 11) is an enzyme that in humans is encoded by the ''MRE11'' gene. The gene has been designated ''MRE11A'' to distinguish it from the pseudogene ''MRE11B'' that is nowadays named ''MRE11P1''. Function This gene encodes a nuclear protein involved in homologous recombination, telomere length maintenance, and DNA double-strand break repair. By itself, the protein has 3' to 5' exonuclease activity and endonuclease activity. The protein forms a complex with the RAD50 homolog; this complex is required for nonhomologous joining of DNA ends and possesses increased single-stranded DNA endonuclease and 3' to 5' exonuclease activities. In conjunction with a DNA ligase, this protein promotes the joining of noncomplementary ends in vitro using short homologies near the ends of the DNA fragments. This gene has a pseudogene on chromosome 3. Alternative splicing of this gene results in two transcript variants encoding different is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neutrophil Cytosolic Factor 4
Neutrophil cytosol factor 4 is a protein that in humans is encoded by the ''NCF4'' gene. Function The protein encoded by this gene is a cytosolic regulatory component of the superoxide-producing phagocyte NADPH-oxidase, a multicomponent enzyme system important for host defense. This protein is preferentially expressed in cells of myeloid lineage. It interacts primarily with neutrophil cytosolic factor 2 (NCF2/p67-phox) to form a complex with neutrophil cytosolic factor 1 (NCF1/p47-phox), which further interacts with the small G protein RAC1 and translocates to the membrane upon cell stimulation. This complex then activates flavocytochrome b, the membrane-integrated catalytic core of the enzyme system. The PX domain of this protein can bind phospholipid products of the PI(3) kinase, which suggests its role in PI(3) kinase-mediated signaling events. The phosphorylation of this protein was found to negatively regulate the enzyme activity. Alternatively spliced transcript variants e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomerase Reverse Transcriptase
Telomerase reverse transcriptase (abbreviated to TERT, or hTERT in humans) is a catalytic subunit of the enzyme telomerase, which, together with the telomerase RNA component (TERC), comprises the most important unit of the telomerase complex. Telomerases are part of a distinct subgroup of RNA-dependent polymerases. Telomerase lengthens telomeres in DNA strands, thereby allowing senescent cells that would otherwise become postmitotic and undergo apoptosis to exceed the Hayflick limit and become potentially immortal, as is often the case with cancerous cells. To be specific, TERT is responsible for catalyzing the addition of nucleotides in a TTAGGG sequence to the ends of a chromosome's telomeres. This addition of repetitive DNA sequences prevents degradation of the chromosomal ends following multiple rounds of replication. hTERT absence (usually as a result of a chromosomal mutation) is associated with the disorder Cri du chat. Function Telomerase is a ribonucleopr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HOXC4
Homeobox protein Hox-C4 is a protein that in humans is encoded by the ''HOXC4'' gene. Function This gene belongs to the homeobox family of genes. The homeobox genes encode a highly conserved family of transcription factors that play an important role in morphogenesis in all multicellular organisms. Mammals possess four similar homeobox gene clusters, HOXA, HOXB, HOXC and HOXD, which are located on different chromosomes and consist of 9 to 11 genes arranged in tandem. This gene, HOXC4, is one of several homeobox HOXC genes located in a cluster on chromosome 12. Three genes, HOXC5, HOXC4 and HOXC6, share a 5' non-coding exon. Transcripts may include the shared exon spliced to the gene-specific exons, or they may include only the gene-specific exons. Two alternatively spliced variants that encode the same protein have been described for HOXC4. Transcript variant one includes the shared exon, and transcript variant two includes only gene-specific exons. See also * Homeobox ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
VAV1
Proto-oncogene vav is a protein that in humans is encoded by the ''VAV1'' gene. Function The protein encoded by this proto-oncogene is a member of the Dbl family of guanine nucleotide exchange factors (GEF) for the Rho family of GTP binding proteins. The protein is important in hematopoiesis, playing a role in T-cell and B-cell development and activation. This particular GEF has been identified as the specific binding partner of Nef proteins from HIV-1. Coexpression and binding of these partners initiates profound morphological changes, cytoskeletal rearrangements and the JNK/SAPK signaling cascade, leading to increased levels of viral transcription and replication. Interactions VAV1 has been shown to interact with: * ARHGDIB, * Abl gene, * Cbl gene * EZH2, * Grb2 Growth factor receptor-bound protein 2, also known as Grb2, is an adaptor protein involved in signal transduction/ cell communication. In humans, the GRB2 protein is encoded by the ''GRB2'' gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GCN5L2
__NOTOC__ Histone acetyltransferase KAT2A is an enzyme that in humans is encoded by the ''KAT2A'' gene. Interactions GCN5L2 has been shown to interact with: * DDB1, * Ku70, * Ku80 Ku80 is a protein that, in humans, is encoded by the ''XRCC5'' gene. Together, Ku70 and Ku80 make up the Ku heterodimer, which binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA rep ..., * TADA2L, * TAF9, and * Transcription initiation protein SPT3 homolog. References Further reading * * * * * * * * * * * * * * * * * * External links * {{gene-17-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |