|
Endoplasmic-reticulum-associated Protein Degradation
Endoplasmic-reticulum-associated protein degradation (ERAD) designates a Cell (biology), cellular pathway which targets misfolded proteins of the endoplasmic reticulum for ubiquitination and subsequent degradation by a protein-degrading complex, called the proteasome. Mechanism The process of ERAD can be divided into three steps: Recognition of misfolded or mutated proteins in the endoplasmic reticulum The recognition of misfolded or mutated proteins depends on the detection of substructures within proteins such as exposed hydrophobic regions, unpaired cysteine residues and immature glycans. In mammalian cells for example, there exists a mechanism called glycan processing. In this mechanism, the lectin-type Chaperone (protein), chaperones calnexin/calreticulin (CNX/CRT) provide immature glycoproteins the opportunity to reach their native conformation. They can do this by way of reglucosylating these glycoproteins by an enzyme called Uridine diphosphate, UDP-glucose-glycopro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UGGT
UGGT, or UDP-glucose:glycoprotein glucosyltransferase, is a soluble enzyme resident in the lumen of the endoplasmic reticulum (ER). The main function of UGGT is to recognize misfolded glycoproteins and transfer a glucose (Glc) monomer (monoglucosylate) to the terminal mannose of the A-branch of the glycan on the glycoprotein. It uses UDP-glucose (UDP-Glc) as the glucosyl donor and requires calcium ions for its activity: misfolded-glycoprotein-Asn-GlcNAc2Man9 + UDP-Glc => misfolded-glycoprotein-Asn-GlcNAc2Man9Glc1 + UDP UGGT is about 170 kDa and it consists of two structurally independent portions: a variable N-terminal portion of ~1200 amino acids, which in turn comprises 4 thioredoxin-like domains and two beta-sandwich domains, and senses glycoprotein misfolding; and a highly conserved C-terminal catalytic portion of ~300 amino acids, folding as a glucosyltransferase domain belonging to fold family GT24. Higher eukaryotes possess two isoforms, UGGT1 and UGGT2, but only in 202 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polypeptide Chain
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty amino acids are called oligopeptides, and include dipeptides, tripeptides, and tetrapeptides. Peptides fall under the broad chemical classes of biological polymers and oligomers, alongside nucleic acids, oligosaccharides, polysaccharides, and others. Proteins consist of one or more polypeptides arranged in a biologically functional way, often bound to ligands such as coenzymes and cofactors, to another protein or other macromolecule such as DNA or RNA, or to complex macromolecular assemblies. Amino acids that have been incorporated into peptides are termed residues. A water molecule is released during formation of each amide bond.. All peptides except cyclic peptides have an N-terminal (amine group) and C-terminal (carboxyl group) res ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex. Large assemblies of proteins such as viruses often use a small number of types of protein subunits as building blocks. A subunit is often named with a Greek or Roman letter, and the numbers of this type of subunit in a protein is indicated by a subscript. For example, ATP synthase has a type of subunit called α. Three of these are present in the ATP synthase molecule, leading to the designation α3. Larger groups of subunits can also be specified, like α3β3-hexamer and c-ring. Naturally occurring proteins that have a relatively small number of subunits are referred to as oligomeric.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-conjugating Enzyme
Ubiquitin-conjugating enzymes, also known as E2 enzymes and more rarely as ''ubiquitin-carrier enzymes'', perform the second step in the ubiquitination reaction that targets a protein for degradation via the proteasome. The ubiquitination process covalently attaches ubiquitin, a short protein of 76 amino acids, to a lysine residue on the target protein. Once a protein has been tagged with one ubiquitin molecule, additional rounds of ubiquitination form a polyubiquitin chain that is recognized by the proteasome's 19S regulatory particle, triggering the adenosine triphosphate, ATP-dependent denaturation (biochemistry), unfolding of the target protein that allows passage into the proteasome's 20S core particle, where proteases degrade the target into short peptide fragments for recycling by the cell (biology), cell. Relationships A ubiquitin-activating enzyme, or E1, first activates the ubiquitin by covalently attaching the molecule to its active site cysteine residue. The activated u ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ... or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is translated from messenger RNA, it is created from N-terminus to C-terminus. The convention for writing peptide sequences is to put the C-terminal end on the right and write the sequence from N- to C-terminus. Chemistry Each amino acid has a carboxyl group and an amine group. Amino acids link to one another to form a chain by a dehydration reaction which joins the amine group of one amino acid to the carboxyl group of the next. Thus polypeptide chains have an end with an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
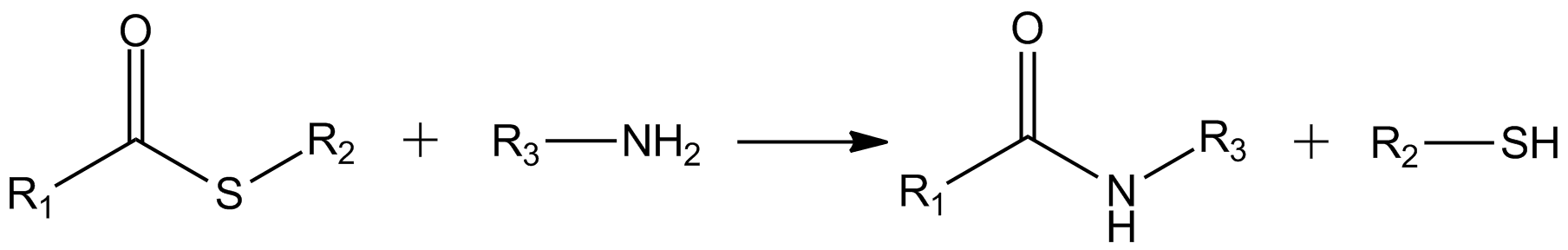
Thioester
In organic chemistry, thioesters are organosulfur compounds with the molecular structure . They are analogous to carboxylate esters () with the sulfur in the thioester replacing oxygen in the carboxylate ester, as implied by the thio- prefix. They are the product of esterification of a carboxylic acid () with a thiol (). In biochemistry, the best-known thioesters are derivatives of coenzyme A, e.g., acetyl-CoA.Matthys J. Janssen "Carboxylic Acids and Esters" in PATAI's Chemistry of Functional Groups: Carboxylic Acids and Esters, Saul Patai, Ed. John Wiley, 1969, New York: pp. 705–764. The R and R' represent organyl groups, or H in the case of R. Synthesis One route to thioesters involves the reaction of an acid chloride with an alkali metal salt of a thiol: : Another common route entails the displacement of halides by the alkali metal salt of a thiocarboxylic acid. For example, thioacetate esters are commonly prepared by alkylation of potassium thioacetate: : Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Triphosphate
Adenosine triphosphate (ATP) is a nucleoside triphosphate that provides energy to drive and support many processes in living cell (biology), cells, such as muscle contraction, nerve impulse propagation, and chemical synthesis. Found in all known forms of life, it is often referred to as the "molecular unit of currency" for intracellular energy transfer. When consumed in a Metabolism, metabolic process, ATP converts either to adenosine diphosphate (ADP) or to adenosine monophosphate (AMP). Other processes regenerate ATP. It is also a Precursor (chemistry), precursor to DNA and RNA, and is used as a coenzyme. An average adult human processes around 50 kilograms (about 100 mole (unit), moles) daily. From the perspective of biochemistry, ATP is classified as a nucleoside triphosphate, which indicates that it consists of three components: a nitrogenous base (adenine), the sugar ribose, and the Polyphosphate, triphosphate. Structure ATP consists of three parts: a sugar, an amine base ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-activating Enzyme
Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which (among other things) can target a protein for degradation via a proteasome. This covalent bond of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms. Many processes such as cell division, immune responses and embryonic development are also regulated by post-translational modification by ubiquitin and ubiquitin-like proteins. Ubiquitination (ubiquitylation) Ubiquitin-activating enzyme (E1) starts the ubiquitination process (Figure 1). The E1 enzyme, along with ATP, binds to the ubiquitin protein. The E1 enzyme then passes the ubiquitin protein to a second protein, called ubiquitin carrier or conjugation protein (E2). The E2 protein complexes with a ubiquitin protein ligase (E3). This ubiquitin protein ligase recognizes which protein needs to be tagged and catalyzes the transfer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymatic
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include catalytic RNA molecules, also called ribozymes. They are sometimes described as a ''type'' of enzyme rather than being ''like'' an enzyme, but even in the decades ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Valosin-containing Protein
Valosin-containing protein (VCP) or transitional endoplasmic reticulum ATPase (TER ATPase) also known as p97 in mammals and CDC48 in '' S. cerevisiae,'' is an enzyme that in humans is encoded by the ''VCP'' gene. The TER ATPase is an ATPase enzyme present in all eukaryotes and archaebacteria. Its main function is to segregate protein molecules from large cellular structures such as protein assemblies, organelle membranes and chromatin, and thus facilitate the degradation of released polypeptides by the multi-subunit protease proteasome. VCP/p97/CDC48 is a member of the AAA+ (extended family of ATPases associated with various cellular activities) ATPase family. Enzymes of this family are found in all species from bacteria to humans. Many of them are important chaperones that regulate folding or unfolding of substrate proteins. VCP is a type II AAA+ ATPase, which means that it contains two tandem ATPase domains (named D1 and D2, respectively) (Figure 1). The two ATPase domai ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substrate (biochemistry)
In chemistry, the term substrate is highly context-dependent. Broadly speaking, it can refer either to a chemical species being observed in a chemical reaction, or to a surface on which other chemical reactions or microscopy are performed. In the former sense, a reagent is added to the ''substrate'' to generate a product through a chemical reaction. The term is used in a similar sense in synthetic and organic chemistry, where the substrate is the chemical of interest that is being modified. In biochemistry, an enzyme substrate is the material upon which an enzyme acts. When referring to Le Chatelier's principle, the substrate is the reagent whose concentration is changed. ;Spontaneous reaction : :*Where S is substrate and P is product. ;Catalysed reaction : :*Where S is substrate, P is product and C is catalyst. In the latter sense, it may refer to a surface on which other chemical reactions are performed or play a supporting role in a variety of spectroscopic and mic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |