Southern Hybridization on:
[Wikipedia]
[Google]
[Amazon]


 A Southern blot is a method used in molecular biology for detection of a specific
A Southern blot is a method used in molecular biology for detection of a specific
OpenWetWare
{{Molecular probes Molecular biology techniques



 A Southern blot is a method used in molecular biology for detection of a specific
A Southern blot is a method used in molecular biology for detection of a specific DNA sequence
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. Th ...
in DNA samples. Southern blotting combines transfer of electrophoresis
Electrophoresis, from Ancient Greek ἤλεκτρον (ḗlektron, "amber") and φόρησις (phórēsis, "the act of bearing"), is the motion of dispersed particles relative to a fluid under the influence of a spatially uniform electric fie ...
-separated DNA fragments to a filter membrane and subsequent fragment detection by probe hybridization
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA of usually 15–10000 nucleotide long which can be radioactively or fluorescently labeled. HP can be used to detect the presence of nucleotide sequences in analyzed RNA ...
.
The method is named after the British biologist
A biologist is a scientist who conducts research in biology. Biologists are interested in studying life on Earth, whether it is an individual cell, a multicellular organism, or a community of interacting populations. They usually specialize in ...
Edwin Southern, who first published it in 1975. Other blotting methods (i.e., western blot, northern blot, eastern blot, southwestern blot) that employ similar principles, but using RNA or protein, have later been named in reference to Edwin Southern's name. As the label is eponymous, Southern is capitalised, as is conventional of proper nouns. The names for other blotting methods may follow this convention, by analogy.
Method
#Restrictionendonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
s are used to cut high-molecular-weight DNA strands into smaller fragments.
#The DNA fragments are then electrophoresed on an agarose gel to separate them by size.
# If some of the DNA fragments are larger than 15 kb, then prior to blotting, the gel may be treated with an acid, such as dilute HCl. This depurinates the DNA fragments, breaking the DNA into smaller pieces, thereby allowing more efficient transfer from the gel to membrane.
# If alkaline transfer methods are used, the DNA gel is placed into an alkaline solution (typically containing sodium hydroxide
Sodium hydroxide, also known as lye and caustic soda, is an inorganic compound with the formula NaOH. It is a white solid ionic compound consisting of sodium cations and hydroxide anions .
Sodium hydroxide is a highly caustic base and alkali ...
) to denature the double-stranded DNA. The denaturation in an alkaline environment may improve binding of the negatively charged thymine residues of DNA to a positively charged amino groups of membrane, separating it into single DNA strands for later hybridization
Hybridization (or hybridisation) may refer to:
*Hybridization (biology), the process of combining different varieties of organisms to create a hybrid
*Orbital hybridization, in chemistry, the mixing of atomic orbitals into new hybrid orbitals
*Nu ...
to the probe (see below), and destroys any residual RNA that may still be present in the DNA. The choice of alkaline over neutral transfer methods, however, is often empirical and may result in equivalent results.
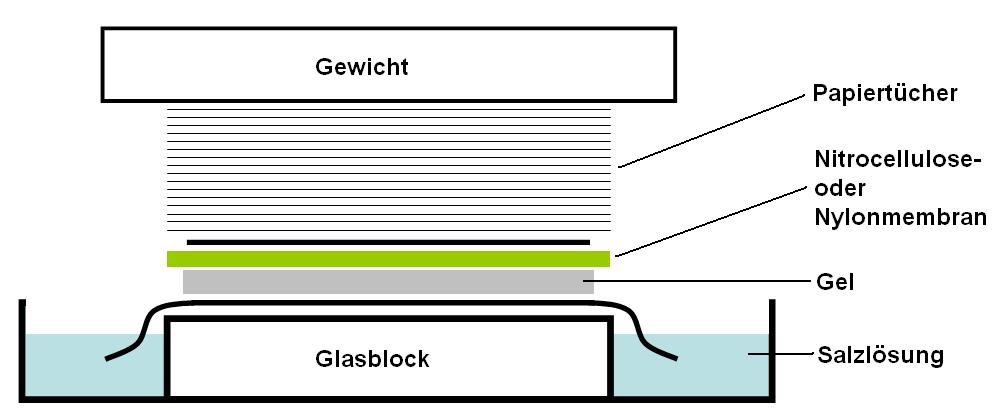
# A sheet of nitrocellulose (or, alternatively, nylon) membrane is placed on top of (or below, depending on the direction of the transfer) the gel. Pressure is applied evenly to the gel (either using suction, or by placing a stack of paper towels and a weight on top of the membrane and gel), to ensure good and even contact between gel and membrane. If transferring by suction, 20X SSC buffer is used to ensure a seal and prevent drying of the gel. Buffer transfer by capillary action from a region of high water potential
Water potential is the potential energy of water per unit volume relative to pure water in reference conditions. Water potential quantifies the tendency of water to move from one area to another due to osmosis, gravity, mechanical pressure and ...
to a region of low water potential (usually filter paper and paper tissues) is then used to move the DNA from the gel onto the membrane; ion exchange interactions bind the DNA to the membrane due to the negative charge of the DNA and positive charge of the membrane.
# The membrane is then baked in a vacuum or regular oven at 80 °C for 2 hours (standard conditions; nitrocellulose or nylon membrane) or exposed to ultraviolet radiation (nylon membrane) to permanently attach the transferred DNA to the membrane.
# The membrane is then exposed to a hybridization probe—a single DNA fragment with a specific sequence whose presence in the target DNA is to be determined. The probe DNA is labelled so that it can be detected, usually by incorporating radioactivity or tagging the molecule with a fluorescent or chromogenic dye. In some cases, the hybridization probe may be made from RNA, rather than DNA. To ensure the specificity of the binding of the probe to the sample DNA, most common hybridization methods use salmon or herring sperm DNA for blocking of the membrane surface and target DNA, deionized formamide, and detergents such as SDS to reduce non-specific binding of the probe.
# After hybridization, excess probe is washed from the membrane (typically using SSC buffer), and the pattern of hybridization is visualized on X-ray film by autoradiography
An autoradiograph is an image on an X-ray film or nuclear emulsion produced by the pattern of decay emissions (e.g., beta particles or gamma rays) from a distribution of a radioactive substance. Alternatively, the autoradiograph is also available ...
in the case of a radioactive or fluorescent probe, or by development of colour on the membrane if a chromogenic detection method is used.
Result
Hybridization of the probe to a specific DNA fragment on the filter membrane indicates that this fragment contains DNA sequence that is complementary to the probe. The transfer step of the DNA from the electrophoresis gel to a membrane permits easy binding of the labeled hybridization probe to the size-fractionated DNA. It also allows for the fixation of the target-probe hybrids, required for analysis byautoradiography
An autoradiograph is an image on an X-ray film or nuclear emulsion produced by the pattern of decay emissions (e.g., beta particles or gamma rays) from a distribution of a radioactive substance. Alternatively, the autoradiograph is also available ...
or other detection methods.
Southern blots performed with restriction enzyme-digested genomic DNA may be used to determine the number of sequences (e.g., gene copies) in a genome. A probe that hybridizes only to a single DNA segment that has not been cut by the restriction enzyme will produce a single band on a Southern blot, whereas multiple bands will likely be observed when the probe hybridizes to several highly similar sequences (e.g., those that may be the result of sequence duplication). Modification of the hybridization conditions (for example, increasing the hybridization temperature or decreasing salt concentration) may be used to increase specificity and decrease hybridization of the probe to sequences that are less than 100% similar.
Applications
Southern blotting transfer may be used for homology-based cloning on the basis of amino acid sequence of the protein product of the target gene. Oligonucleotides are designed so that they are complementary to the target sequence. The oligonucleotides are chemically synthesized, radiolabeled, and used to screen aDNA library
In molecular biology, a library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning. There are different types of DNA libraries, including cDNA libraries ( ...
, or other collections of cloned DNA fragments. Sequences that hybridize with the hybridization probe are further analysed, for example, to obtain the full length sequence of the targeted gene.
Southern blotting can also be used to identify methylated sites in particular genes. Particularly useful are the restriction nucleases ''MspI'' and ''HpaII'', both of which recognize and cleave within the same sequence. However, ''HpaII'' requires that a C within that site be methylated, whereas ''MspI'' cleaves only DNA unmethylated at that site. Therefore, any methylated sites within a sequence analyzed with a particular probe will be cleaved by the former, but not the latter, enzyme.Biochemistry 3rd Edition, Matthews, Van Holde et al, Addison Wesley Publishing, pg 977
See also
* Gel electrophoresis of nucleic acids *Restriction fragment
A restriction fragment is a DNA fragment resulting from the cutting of a DNA strand by a restriction enzyme (restriction endonucleases), a process called restriction. Each restriction enzyme is highly specific, recognising a particular short DNA s ...
* Genetic fingerprint
* Northern blot
* Western blot
* Eastern blot
* Southwestern blot
*Northwestern blot The northwestern blot, also known as the northwestern assay, is a hybrid analytical technique of the western blot and the northern blot, and is used in molecular biology to detect interactions between RNA and proteins. A related technique, the west ...
References
External links
OpenWetWare
{{Molecular probes Molecular biology techniques