|
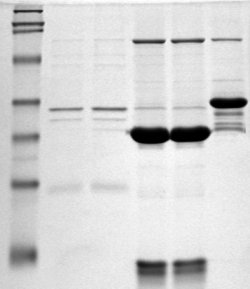
Southwestern Blot
The southwestern blot, is a lab technique that involves identifying as well as characterizing DNA-binding proteins by their ability to bind to specific oligonucleotide probes. Determination of molecular weight of proteins binding to DNA is also made possible by the technique. The name originates from a combination of ideas underlying Southern blotting and Western blotting techniques of which they detect DNA and protein respectively. Similar to other types of blotting, proteins are separated by SDS-PAGE and are subsequently transferred to nitrocellulose membranes. Thereafter southwestern blotting begins to vary with regards to procedure as since the first blotting’s, many more have been proposed and discovered with goals of enhancing results. Former protocols were hampered by the need for large amounts of proteins and their susceptibility to degradation while being isolated. Southwestern blotting was first described by Brian Bowen, Jay Steinberg, U.K. Laemmli, and Harold Weint ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Blotting Compass For Molecular Probes
In molecular biology and genetics, a blot is a method of transferring large biomolecules (proteins, DNA or RNA) onto a carrier, such as a membrane composed of nitrocellulose, polyvinylidene fluoride or nylon. In many instances, this is done after a gel electrophoresis, transferring the molecules from the gel onto the Blotting matrix, blotting membrane, and other times adding the samples directly onto the membrane. After the blotting, the transferred molecules are then visualized by colorant staining (for example, silver staining of proteins), autoradiographic visualization of radioactive tracer, radiolabelled molecules (performed before the blot), or specific labelling of some proteins or nucleic acids. The latter is done with antibody, antibodies or hybridization probes that bind only to some molecules of the blot and have an enzyme joined to them. After proper washing, this enzymatic activity (and so, the molecules found in the blot) is visualized by incubation with a proper reag ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polyacrylamide Gel Electrophoresis
Polyacrylamide gel electrophoresis (PAGE) is a technique widely used in biochemistry, forensic chemistry, genetics, molecular biology and biotechnology to separate biological macromolecules, usually proteins or nucleic acids, according to their Electrophoresis, electrophoretic mobility. Electrophoretic mobility is a function of the length, conformation, and charge of the molecule. Polyacrylamide gel electrophoresis is a powerful tool used to analyze RNA samples. When polyacrylamide gel is denatured after electrophoresis, it provides information on the sample composition of the RNA species. Hydration reaction, Hydration of acrylonitrile results in formation of acrylamide molecules () by nitrile hydratase. Acrylamide monomer is in a powder state before addition of water. Acrylamide is toxic to the human nervous system, therefore all safety measures must be followed when working with it. Acrylamide is soluble in water and upon addition of free-radical initiators it polymerizes resu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sodium Dodecyl Sulfate
Sodium dodecyl sulfate (SDS) or sodium lauryl sulfate (SLS), sometimes written sodium laurilsulfate, is an organic compound with the formula and structure . It is an anionic surfactant used in many cleaning and hygiene products. This compound is the sodium salt of the 12-carbon organosulfate. Its hydrocarbon tail combined with a polar " headgroup" give the compound amphiphilic properties that make it useful as a detergent. SDS is also component of mixtures produced from inexpensive coconut and palm oils. SDS is a common component of many domestic cleaning, personal hygiene and cosmetic, pharmaceutical, and food products, as well as of industrial and commercial cleaning and product formulations. Physicochemical properties The critical micelle concentration (CMC) in water at 25 °C is 8.2 mM, and the aggregation number at this concentration is usually considered to be about 62. The micelle ionization fraction (α) is around 0.3 (or 30%). Applications Cleaning and hygie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Two-dimensional Gel Electrophoresis
A two-dimensional space is a mathematical space with two dimensions, meaning points have two degrees of freedom: their locations can be locally described with two coordinates or they can move in two independent directions. Common two-dimensional spaces are often called '' planes'', or, more generally, '' surfaces''. These include analogs to physical spaces, like flat planes, and curved surfaces like spheres, cylinders, and cones, which can be infinite or finite. Some two-dimensional mathematical spaces are not used to represent physical positions, like an affine plane or complex plane. Flat The most basic example is the flat Euclidean plane, an idealization of a flat surface in physical space such as a sheet of paper or a chalkboard. On the Euclidean plane, any two points can be joined by a unique straight line along which the distance can be measured. The space is flat because any two lines transversed by a third line perpendicular to both of them are parallel, meaning th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mass Spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used in many different fields and is applied to pure samples as well as complex mixtures. A mass spectrum is a type of plot of the ion signal as a function of the mass-to-charge ratio. These spectra are used to determine the elemental or isotopic signature of a sample, the masses of particles and of molecules, and to elucidate the chemical identity or structure of molecules and other chemical compounds. In a typical MS procedure, a sample, which may be solid, liquid, or gaseous, is ionization, ionized, for example by bombarding it with a Electron ionization, beam of electrons. This may cause some of the sample's molecules to break up into positively charged fragments or simply become positively charged without fragmenting. These ions (fragmen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
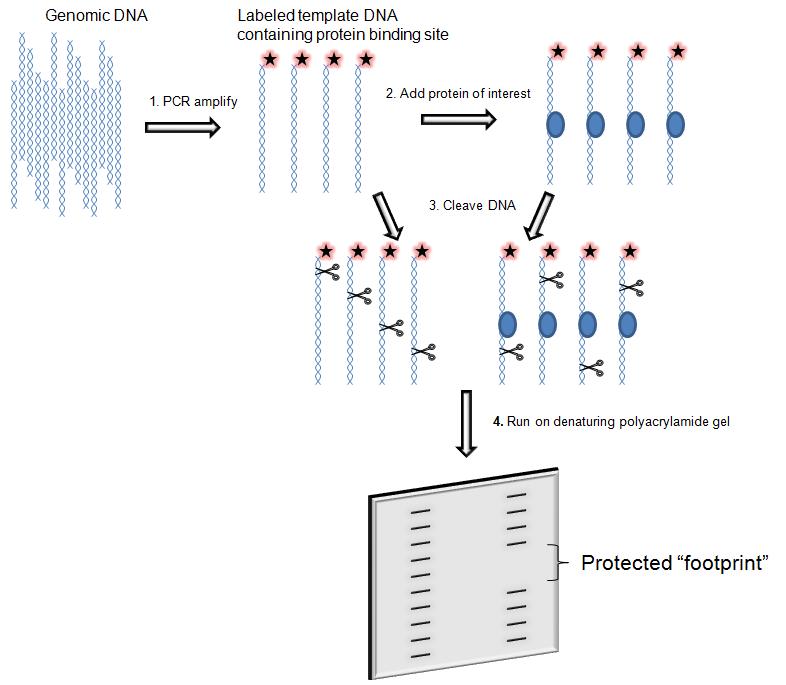
DNA Footprinting
DNA footprinting is a method of ''in vitro'' DNA analysis that assists researchers in determining transcription factor (TF) associated binding proteins. This technique can be used to study protein-DNA interactions both outside and within cells. Transcription factors are regulatory proteins that assist with various levels of DNA regulation. These regulatory molecules and associated proteins bind Promoter (biology), promoters, enhancers, or Silencer (DNA), silencers to drive or repress transcription and are fundamental to understanding the unique regulation of individual genes within the genome. First developed in 1978, primary investigators David J. Galas, Ph.D. and Albert Schmitz, Ph.D. modified the pre-existing Maxam–Gilbert sequencing, Maxam-Gilbert chemical sequencing technique to bind specifically to the lac repressor protein. Since the technique's discovery, scientific researchers have developed this technique to map chromatin and have greatly reduced technical requirements ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bovine Serum Albumin
Bovine serum albumin (BSA or "Fraction V") is a serum albumin protein derived from cows. It is often used as a protein concentration standard in lab experiments. The nickname "Fraction V" refers to albumin being the fifth fraction of the original Edwin Cohn purification methodology that made use of differential solubility characteristics of plasma proteins. By manipulating solvent concentrations, pH, salt levels, and temperature, Cohn was able to pull out successive "fractions" of blood plasma. The process was first commercialized with human albumin for medical use and later adopted for production of BSA. Properties The full-length BSA precursor polypeptide is 607 amino acids (AAs) in length. An N-terminal 18-residue signal peptide is cut off from the precursor protein upon secretion, hence the initial protein product contains 589 amino acid residues. An additional six amino acids are cleaved to yield the mature BSA protein that contains 583 amino acids. BSA has thre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Denaturation (biochemistry)
In biochemistry, denaturation is a process in which proteins or nucleic acids lose folded structure present in their native state due to various factors, including application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent (e.g., alcohol or chloroform), agitation, radiation, or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility or dissociation of cofactors to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called ''coagulation''. Denatured proteins, e.g., metalloenzymes, lose their 3D structure or metal cofactor, and therefore, cannot function. Proper protein folding is key to whether a globular or memb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electroblotting
Electroblotting is a method in molecular biology/biochemistry/immunogenetics to transfer proteins or nucleic acids onto a membrane by using PVDF or nitrocellulose, after gel electrophoresis. The protein or nucleic acid can then be further analyzed using probes such as specific antibodies, ligands like lectins, or stains. This method can be used with all polyacrylamide and agarose gels. An alternative technique for transferring proteins from a gel is capillary blotting. Development This technique was patented in 1989 by William J. Littlehales under the title "Electroblotting technique for transferring specimens from a polyacrylamide electrophoresis or like gel onto a membrane. Electroblotting procedure This technique relies upon current and a transfer buffer solution to drive proteins or nucleic acids onto a membrane. Following electrophoresis, a standard tank or semi-dry blotting transfer system is set up. A stack is put together in the following order from cathode to anode: spon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA enzyme substrate (biology), substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each backbone chain, sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |