full genome sequencing on:
[Wikipedia]
[Google]
[Amazon]
 Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the
Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the

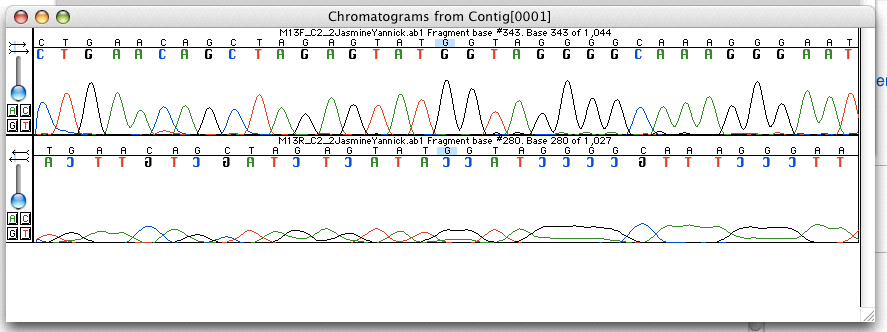
 The DNA sequencing methods used in the 1970s and 1980s were manual; for example, Maxam–Gilbert sequencing and Sanger sequencing. Several whole bacteriophage and animal viral genomes were sequenced by these techniques, but the shift to more rapid, automated sequencing methods in the 1990s facilitated the sequencing of the larger bacterial and eukaryotic genomes.
The first virus to have its complete genome sequenced was the Bacteriophage MS2 by 1976. In 1992, yeast chromosome III was the first chromosome of any organism to be fully sequenced. The first organism whose entire genome was fully sequenced was ''
The DNA sequencing methods used in the 1970s and 1980s were manual; for example, Maxam–Gilbert sequencing and Sanger sequencing. Several whole bacteriophage and animal viral genomes were sequenced by these techniques, but the shift to more rapid, automated sequencing methods in the 1990s facilitated the sequencing of the larger bacterial and eukaryotic genomes.
The first virus to have its complete genome sequenced was the Bacteriophage MS2 by 1976. In 1992, yeast chromosome III was the first chromosome of any organism to be fully sequenced. The first organism whose entire genome was fully sequenced was ''
 Sequencing of nearly an entire human genome was first accomplished in 2000 partly through the use of shotgun sequencing technology. While full genome shotgun sequencing for small (4000–7000
Sequencing of nearly an entire human genome was first accomplished in 2000 partly through the use of shotgun sequencing technology. While full genome shotgun sequencing for small (4000–7000
 A number of public and private companies are competing to develop a full genome sequencing platform that is commercially robust for both research and clinical use, including Illumina, Knome, Sequenom,
454 Life Sciences, Pacific Biosciences, Complete Genomics,
Helicos Biosciences, GE Global Research (
A number of public and private companies are competing to develop a full genome sequencing platform that is commercially robust for both research and clinical use, including Illumina, Knome, Sequenom,
454 Life Sciences, Pacific Biosciences, Complete Genomics,
Helicos Biosciences, GE Global Research (
James Watson's Personal Genome Sequence
AAAS/Science: Genome Sequencing Poster
{{Breakthrough of the Year Molecular biology DNA sequencing Biotechnology Genomics Bioinformatics DNA * Gene tests Molecular genetics

 Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the
Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
sequence of an organism's genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is us ...
and, for plants, in the chloroplast
A chloroplast () is a type of membrane-bound organelle, organelle known as a plastid that conducts photosynthesis mostly in plant cell, plant and algae, algal cells. Chloroplasts have a high concentration of chlorophyll pigments which captur ...
.
Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of gene sequencing at SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology
Evolutionary biology is the subfield of biology that studies the evolutionary processes such as natural selection, common descent, and speciation that produced the diversity of life on Earth. In the 1930s, the discipline of evolutionary biolo ...
, and hence may lay the foundation for predicting disease susceptibility and drug response.
Whole genome sequencing should not be confused with DNA profiling
DNA profiling (also called DNA fingerprinting and genetic fingerprinting) is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is cal ...
, which only determines the likelihood that genetic material came from a particular individual or group, and does not contain additional information on genetic relationships, origin or susceptibility to specific diseases. In addition, whole genome sequencing should not be confused with methods that sequence specific subsets of the genome – such methods include whole exome sequencing (1–2% of the genome) or SNP genotyping (< 0.1% of the genome).
History


 The DNA sequencing methods used in the 1970s and 1980s were manual; for example, Maxam–Gilbert sequencing and Sanger sequencing. Several whole bacteriophage and animal viral genomes were sequenced by these techniques, but the shift to more rapid, automated sequencing methods in the 1990s facilitated the sequencing of the larger bacterial and eukaryotic genomes.
The first virus to have its complete genome sequenced was the Bacteriophage MS2 by 1976. In 1992, yeast chromosome III was the first chromosome of any organism to be fully sequenced. The first organism whose entire genome was fully sequenced was ''
The DNA sequencing methods used in the 1970s and 1980s were manual; for example, Maxam–Gilbert sequencing and Sanger sequencing. Several whole bacteriophage and animal viral genomes were sequenced by these techniques, but the shift to more rapid, automated sequencing methods in the 1990s facilitated the sequencing of the larger bacterial and eukaryotic genomes.
The first virus to have its complete genome sequenced was the Bacteriophage MS2 by 1976. In 1992, yeast chromosome III was the first chromosome of any organism to be fully sequenced. The first organism whose entire genome was fully sequenced was ''Haemophilus influenzae
''Haemophilus influenzae'' (formerly called Pfeiffer's bacillus or ''Bacillus influenzae'') is a Gram-negative, Motility, non-motile, Coccobacillus, coccobacillary, facultative anaerobic organism, facultatively anaerobic, Capnophile, capnophili ...
'' in 1995. After it, the genomes of other bacteria and some archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
were first sequenced, largely due to their small genome size. ''H. influenzae'' has a genome of 1,830,140 base pairs of DNA. In contrast, eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
, both unicellular
A unicellular organism, also known as a single-celled organism, is an organism that consists of a single cell, unlike a multicellular organism that consists of multiple cells. Organisms fall into two general categories: prokaryotic organisms and ...
and multicellular
A multicellular organism is an organism that consists of more than one cell (biology), cell, unlike unicellular organisms. All species of animals, Embryophyte, land plants and most fungi are multicellular, as are many algae, whereas a few organism ...
such as '' Amoeba dubia'' and humans (''Homo sapiens
Humans (''Homo sapiens'') or modern humans are the most common and widespread species of primate, and the last surviving species of the genus ''Homo''. They are Hominidae, great apes characterized by their Prehistory of nakedness and clothing ...
'') respectively, have much larger genomes (see C-value paradox). ''Amoeba dubia'' has a genome of 700 billion nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
pairs spread across thousands of chromosomes. Humans contain fewer nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
pairs (about 3.2 billion in each germ cell
A germ cell is any cell that gives rise to the gametes of an organism that reproduces sexually. In many animals, the germ cells originate in the primitive streak and migrate via the gut of an embryo to the developing gonads. There, they unde ...
– note the exact size of the human genome is still being revised) than ''A. dubia,'' however, their genome size far outweighs the genome size of individual bacteria.
The first bacterial and archaeal genomes, including that of ''H. influenzae'', were sequenced by Shotgun sequencing. In 1996, the first eukaryotic genome (''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
'') was sequenced. ''S. cerevisiae'', a model organism
A model organism is a non-human species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the model organism will provide insight into the workings of other organisms. Mo ...
in biology
Biology is the scientific study of life and living organisms. It is a broad natural science that encompasses a wide range of fields and unifying principles that explain the structure, function, growth, History of life, origin, evolution, and ...
has a genome of only around 12 million nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
pairs, and was the first ''unicellular'' eukaryote to have its whole genome sequenced. The first ''multicellular'' eukaryote, and animal
Animals are multicellular, eukaryotic organisms in the Biology, biological Kingdom (biology), kingdom Animalia (). With few exceptions, animals heterotroph, consume organic material, Cellular respiration#Aerobic respiration, breathe oxygen, ...
, to have its whole genome sequenced was the nematode
The nematodes ( or ; ; ), roundworms or eelworms constitute the phylum Nematoda. Species in the phylum inhabit a broad range of environments. Most species are free-living, feeding on microorganisms, but many are parasitic. Parasitic worms (h ...
worm: ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a Hybrid word, blend of the Greek ''caeno-'' (recent), ''r ...
'' in 1998. Eukaryotic genomes are sequenced by several methods including Shotgun sequencing of short DNA fragments and sequencing of larger DNA clones from DNA libraries such as bacterial artificial chromosomes (BACs) and yeast artificial chromosomes (YACs).
In 1999, the entire DNA sequence of human chromosome 22, the second shortest human autosome
An autosome is any chromosome that is not a sex chromosome. The members of an autosome pair in a diploid cell have the same morphology, unlike those in allosomal (sex chromosome) pairs, which may have different structures. The DNA in autosomes ...
, was published. By the year 2000, the second animal and second invertebrate
Invertebrates are animals that neither develop nor retain a vertebral column (commonly known as a ''spine'' or ''backbone''), which evolved from the notochord. It is a paraphyletic grouping including all animals excluding the chordata, chordate s ...
(yet first insect
Insects (from Latin ') are Hexapoda, hexapod invertebrates of the class (biology), class Insecta. They are the largest group within the arthropod phylum. Insects have a chitinous exoskeleton, a three-part body (Insect morphology#Head, head, ...
) genome was sequenced – that of the fruit fly ''Drosophila melanogaster
''Drosophila melanogaster'' is a species of fly (an insect of the Order (biology), order Diptera) in the family Drosophilidae. The species is often referred to as the fruit fly or lesser fruit fly, or less commonly the "vinegar fly", "pomace fly" ...
'' – a popular choice of model organism in experimental research. The first plant
Plants are the eukaryotes that form the Kingdom (biology), kingdom Plantae; they are predominantly Photosynthesis, photosynthetic. This means that they obtain their energy from sunlight, using chloroplasts derived from endosymbiosis with c ...
genome – that of the model organism ''Arabidopsis thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally ...
'' – was also fully sequenced by 2000. By 2001, a draft of the entire human genome sequence was published. The genome of the laboratory mouse ''Mus musculus
The house mouse (''Mus musculus'') is a small mammal of the rodent family Muridae, characteristically having a pointed snout, large rounded ears, and a long and almost hairless tail. It is one of the most abundant species of the genus ''Mus (genu ...
'' was completed in 2002.
In 2004, the Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
published an incomplete version of the human genome. In 2008, a group from Leiden, the Netherlands, reported the sequencing of the first female human genome ( Marjolein Kriek).
Currently thousands of genomes have been wholly or partially sequenced.
Experimental details
Cells used for sequencing
Almost any biological sample containing a full copy of the DNA—even a very small amount of DNA orancient DNA
Ancient DNA (aDNA) is DNA isolated from ancient sources (typically Biological specimen, specimens, but also environmental DNA). Due to degradation processes (including Crosslinking of DNA, cross-linking, deamination and DNA fragmentation, fragme ...
—can provide the genetic material necessary for full genome sequencing. Such samples may include saliva
Saliva (commonly referred as spit or drool) is an extracellular fluid produced and secreted by salivary glands in the mouth. In humans, saliva is around 99% water, plus electrolytes, mucus, white blood cells, epithelial cells (from which ...
, epithelial cells, bone marrow
Bone marrow is a semi-solid biological tissue, tissue found within the Spongy bone, spongy (also known as cancellous) portions of bones. In birds and mammals, bone marrow is the primary site of new blood cell production (or haematopoiesis). It i ...
, hair
Hair is a protein filament that grows from follicles found in the dermis. Hair is one of the defining characteristics of mammals.
The human body, apart from areas of glabrous skin, is covered in follicles which produce thick terminal and ...
(as long as the hair contains a hair follicle
The hair follicle is an organ found in mammalian skin. It resides in the dermal layer of the skin and is made up of 20 different cell types, each with distinct functions. The hair follicle regulates hair growth via a complex interaction betwee ...
), seed
In botany, a seed is a plant structure containing an embryo and stored nutrients in a protective coat called a ''testa''. More generally, the term "seed" means anything that can be Sowing, sown, which may include seed and husk or tuber. Seeds ...
s, plant leaves, or anything else that has DNA-containing cells.
The genome sequence of a single cell selected from a mixed population of cells can be determined using techniques of ''single cell genome sequencing''. This has important advantages in environmental microbiology in cases where a single cell of a particular microorganism species can be isolated from a mixed population by microscopy on the basis of its morphological or other distinguishing characteristics. In such cases the normally necessary steps of isolation and growth of the organism in culture may be omitted, thus allowing the sequencing of a much greater spectrum of organism genomes.
Single cell genome sequencing is being tested as a method of preimplantation genetic diagnosis, wherein a cell from the embryo created by in vitro fertilization
In vitro fertilisation (IVF) is a process of fertilisation in which an egg is combined with sperm in vitro ("in glass"). The process involves monitoring and stimulating the ovulatory process, then removing an ovum or ova (egg or eggs) from ...
is taken and analyzed before embryo transfer
Embryo transfer refers to a step in the process of assisted reproduction in which embryos are placed into the uterus of a female with the intent to establish a pregnancy. This technique - which is often used in connection with in vitro fertili ...
into the uterus. After implantation, cell-free fetal DNA can be taken by simple venipuncture
In medicine, venipuncture or venepuncture is the process of obtaining intravenous access for the purpose of venous Sampling (medicine)#blood, blood sampling (also called ''phlebotomy'') or intravenous therapy. In healthcare, this procedure is p ...
from the mother and used for whole genome sequencing of the fetus.
Early techniques
 Sequencing of nearly an entire human genome was first accomplished in 2000 partly through the use of shotgun sequencing technology. While full genome shotgun sequencing for small (4000–7000
Sequencing of nearly an entire human genome was first accomplished in 2000 partly through the use of shotgun sequencing technology. While full genome shotgun sequencing for small (4000–7000 base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
) genomes was already in use in 1979, broader application benefited from pairwise end sequencing, known colloquially as ''double-barrel shotgun sequencing''. As sequencing projects began to take on longer and more complicated genomes, multiple groups began to realize that useful information could be obtained by sequencing both ends of a fragment of DNA. Although sequencing both ends of the same fragment and keeping track of the paired data was more cumbersome than sequencing a single end of two distinct fragments, the knowledge that the two sequences were oriented in opposite directions and were about the length of a fragment apart from each other was valuable in reconstructing the sequence of the original target fragment.
The first published description of the use of paired ends was in 1990 as part of the sequencing of the human HPRT locus, although the use of paired ends was limited to closing gaps after the application of a traditional shotgun sequencing approach. The first theoretical description of a pure pairwise end sequencing strategy, assuming fragments of constant length, was in 1991. In 1995, the innovation of using fragments of varying sizes was introduced, and demonstrated that a pure pairwise end-sequencing strategy would be possible on large targets. The strategy was subsequently adopted by The Institute for Genomic Research (TIGR) to sequence the entire genome of the bacterium ''Haemophilus influenzae
''Haemophilus influenzae'' (formerly called Pfeiffer's bacillus or ''Bacillus influenzae'') is a Gram-negative, Motility, non-motile, Coccobacillus, coccobacillary, facultative anaerobic organism, facultatively anaerobic, Capnophile, capnophili ...
'' in 1995, and then by Celera Genomics to sequence the entire fruit fly genome in 2000, and subsequently the entire human genome. Applied Biosystems, now called Life Technologies, manufactured the automated capillary sequencers utilized by both Celera Genomics and The Human Genome Project.
Current techniques
While capillary sequencing was the first approach to successfully sequence a nearly full human genome, it is still too expensive and takes too long for commercial purposes. Since 2005, capillary sequencing has been progressively displaced by high-throughput (formerly "next-generation") sequencing technologies such as Illumina dye sequencing,pyrosequencing
Pyrosequencing is a method of DNA sequencing (determining the order of nucleotides in DNA) based on the "sequencing by synthesis" principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase. Pyrosequ ...
, and SMRT sequencing. All of these technologies continue to employ the basic shotgun strategy, namely, parallelization and template generation via genome fragmentation.
Other technologies have emerged, including Nanopore technology. Though the sequencing accuracy of Nanopore technology is lower than those above, its read length is on average much longer. This generation of long reads is valuable especially in ''de novo'' whole-genome sequencing applications.
Analysis
In principle, full genome sequencing can provide the rawnucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
sequence of an individual organism's DNA at a single point in time. However, further analysis must be performed to provide the biological or medical meaning of this sequence, such as how this knowledge can be used to help prevent disease. Methods for analyzing sequencing data are being developed and refined.
Because sequencing generates a lot of data (for example, there are approximately six billion base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s in each human diploid genome), its output is stored electronically and requires a large amount of computing power and storage capacity.
While analysis of WGS data can be slow, it is possible to speed up this step by using dedicated hardware.
Commercialization
General Electric
General Electric Company (GE) was an American Multinational corporation, multinational Conglomerate (company), conglomerate founded in 1892, incorporated in the New York (state), state of New York and headquartered in Boston.
Over the year ...
), Affymetrix, IBM
International Business Machines Corporation (using the trademark IBM), nicknamed Big Blue, is an American Multinational corporation, multinational technology company headquartered in Armonk, New York, and present in over 175 countries. It is ...
, Intelligent Bio-Systems, Life Technologies, Oxford Nanopore Technologies, and the Beijing Genomics Institute
BGI Group, formerly Beijing Genomics Institute, is a Chinese genomics company with headquarters in Yantian, Shenzhen, Yantian, Shenzhen. The company was originally formed in 1999 as a genetics research center to participate in the Human Genome Pro ...
. These companies are heavily financed and backed by venture capitalists, hedge funds
A hedge fund is a pooled investment fund that holds liquid assets and that makes use of complex trading and risk management techniques to aim to improve investment performance and insulate returns from market risk. Among these portfolio techniq ...
, and investment banks.
A commonly-referenced commercial target for sequencing cost until the late 2010s was $1,000USD, however, the private companies are working to reach a new target of only $100.
Incentive
In October 2006, the X Prize Foundation, working in collaboration with the J. Craig Venter Science Foundation, established theArchon X Prize
The Archon Genomics X PRIZE presented by Express Scripts for Genomics, the second X Prize offered by the X Prize Foundation, based in Playa Vista, California, was announced on October 4, 2006 stating that the prize of "$10 million will be awarded ...
for Genomics, intending to award $10 million to "the first team that can build a device and use it to sequence 100 human genomes within 10 days or less, with an accuracy of no more than one error in every 1,000,000 bases sequenced, with sequences accurately covering at least 98% of the genome, and at a recurring cost of no more than $1,000 per genome". The Archon X Prize
The Archon Genomics X PRIZE presented by Express Scripts for Genomics, the second X Prize offered by the X Prize Foundation, based in Playa Vista, California, was announced on October 4, 2006 stating that the prize of "$10 million will be awarded ...
for Genomics was cancelled in 2013, before its official start date.
History
In 2007, Applied Biosystems started selling a new type of sequencer called SOLiD System. The technology allowed users to sequence 60 gigabases per run. In June 2009, Illumina announced that they were launching their own Personal Full Genome Sequencing Service at a depth of 30× for $48,000 per genome. In August, the founder of Helicos Biosciences, Stephen Quake, stated that using the company's Single Molecule Sequencer he sequenced his own full genome for less than $50,000. In November, Complete Genomics published a peer-reviewed paper in ''Science'' demonstrating its ability to sequence a complete human genome for $1,700. In May 2011, Illumina lowered its Full Genome Sequencing service to $5,000 per human genome, or $4,000 if ordering 50 or more. Helicos Biosciences, Pacific Biosciences, Complete Genomics, Illumina, Sequenom, ION Torrent Systems, Halcyon Molecular, NABsys, IBM, and GE Global appear to all be going head to head in the race to commercialize full genome sequencing. With sequencing costs declining, a number of companies began claiming that their equipment would soon achieve the $1,000 genome: these companies included Life Technologies in January 2012, Oxford Nanopore Technologies in February 2012, and Illumina in February 2014. In 2015, the NHGRI estimated the cost of obtaining a whole-genome sequence at around $1,500. In 2016, Veritas Genetics began selling whole genome sequencing, including a report as to some of the information in the sequencing for $999. In summer 2019, Veritas Genetics cut the cost for WGS to $599. In 2017, BGI began offering WGS for $600. However, in 2015, some noted that effective use of whole gene sequencing can cost considerably more than $1000. Also, reportedly there remain parts of the human genome that have not been fully sequenced by 2017.Comparison with other technologies
DNA microarrays
Full genome sequencing provides information on a genome that is orders of magnitude larger than by DNA arrays, the previous leader in genotyping technology. For humans, DNA arrays currently provide genotypic information on up to one million genetic variants, while full genome sequencing will provide information on all six billion bases in the human genome, or 3,000 times more data. Because of this, full genome sequencing is considered a disruptive innovation to the DNA array markets as the accuracy of both range from 99.98% to 99.999% (in non-repetitive DNA regions) and their consumables cost of $5000 per 6 billion base pairs is competitive (for some applications) with DNA arrays ($500 per 1 million basepairs).Applications
Mutation frequencies
Whole genome sequencing has established themutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
frequency for whole human genomes. The mutation frequency in the whole genome between generations for humans (parent to child) is about 70 new mutations per generation. An even lower level of variation was found comparing whole genome sequencing in blood cells for a pair of monozygotic (identical twins) 100-year-old centenarians. Only 8 somatic differences were found, though somatic variation occurring in less than 20% of blood cells would be undetected.
In the specifically protein coding regions of the human genome, it is estimated that there are about 0.35 mutations that would change the protein sequence between parent/child generations (less than one mutated protein per generation).
In cancer, mutation frequencies are much higher, due to genome instability
Genome instability (also genetic instability or genomic instability) refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or ...
. This frequency can further depend on patient age, exposure to DNA damaging agents (such as UV-irradiation or components of tobacco smoke) and the activity/inactivity of DNA repair mechanisms. Furthermore, mutation frequency can vary between cancer types: in germline cells, mutation rates occur at approximately 0.023 mutations per megabase, but this number is much higher in breast cancer (1.18-1.66 somatic mutations per Mb), in lung cancer (17.7) or in melanomas (≈33). Since the haploid human genome consists of approximately 3,200 megabases, this translates into about 74 mutations (mostly in noncoding regions) in germline DNA per generation, but 3,776-5,312 somatic mutations per haploid genome in breast cancer, 56,640 in lung cancer and 105,600 in melanomas.
The distribution of somatic mutations across the human genome is very uneven, such that the gene-rich, early-replicating regions receive fewer mutations than gene-poor, late-replicating heterochromatin, likely due to differential DNA repair activity. In particular, the histone modification H3K9me3 is associated with high, and H3K36me3 with low mutation frequencies.
Genome-wide association studies
In research, whole-genome sequencing can be used in a Genome-Wide Association Study (GWAS) – a project aiming to determine the genetic variant or variants associated with a disease or some other phenotype.Diagnostic use
In 2009, Illumina released its first whole genome sequencers that were approved for clinical as opposed to research-only use and doctors atacademic medical center
The Academic Medical Center (Dutch: ''Academisch Medisch Centrum''), or AMC, was the university hospital affiliated with the University of Amsterdam. After merging with the VU University Medical Center, it now operates as the Amsterdam Universi ...
s began quietly using them to try to diagnose what was wrong with people whom standard approaches had failed to help. In 2009, a team from Stanford led by Euan Ashley performed clinical interpretation of a full human genome, that of bioengineer Stephen Quake. In 2010, Ashley's team reported whole genome molecular autopsy and in 2011, extended the interpretation framework to a fully sequenced family, the West family, who were the first family to be sequenced on the Illumina platform. The price to sequence a genome at that time was $19,500USD, which was billed to the patient but usually paid for out of a research grant; one person at that time had applied for reimbursement from their insurance company. For example, one child had needed around 100 surgeries by the time he was three years old, and his doctor turned to whole genome sequencing to determine the problem; it took a team of around 30 people that included 12 bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
experts, three sequencing technicians, five physicians, two genetic counsellors and two ethicists to identify a rare mutation in the XIAP that was causing widespread problems.
Due to recent cost reductions (see above) whole genome sequencing has become a realistic application in DNA diagnostics. In 2013, the 3Gb-TEST consortium obtained funding from the European Union to prepare the health care system for these innovations in DNA diagnostics. Quality assessment schemes, Health technology assessment and guidelines have to be in place. The 3Gb-TEST consortium has identified the analysis and interpretation of sequence data as the most complicated step in the diagnostic process. At the Consortium meeting in Athens in September 2014, the Consortium coined the word ''genotranslation'' for this crucial step. This step leads to a so-called ''genoreport''. Guidelines are needed to determine the required content of these reports.
Genomes2People (G2P), an initiative of Brigham and Women's Hospital and Harvard Medical School
Harvard Medical School (HMS) is the medical school of Harvard University and is located in the Longwood Medical and Academic Area, Longwood Medical Area in Boston, Massachusetts. Founded in 1782, HMS is the third oldest medical school in the Un ...
was created in 2011 to examine the integration of genomic sequencing into clinical care of adults and children. G2P's director, Robert C. Green, had previously led the REVEAL study — Risk EValuation and Education for Alzheimer's Disease – a series of clinical trials exploring patient reactions to the knowledge of their genetic risk for Alzheimer's.
In 2018, researchers at Rady Children's Hospital Institute for Genomic Medicine in San Diego
San Diego ( , ) is a city on the Pacific coast of Southern California, adjacent to the Mexico–United States border. With a population of over 1.4 million, it is the List of United States cities by population, eighth-most populous city in t ...
determined that rapid whole-genome sequencing (rWGS) could diagnose genetic disorders in time to change acute medical or surgical management (clinical utility) and improve outcomes in acutely ill infants. In a retrospective cohort study of acutely ill inpatient infants in a regional children's hospital from July 2016-March 2017, forty-two families received rWGS for etiologic diagnosis of genetic disorders. The diagnostic sensitivity of rWGS was 43% (eighteen of 42 infants) and 10% (four of 42 infants) for standard genetic tests (P = .0005). The rate of clinical utility of rWGS (31%, thirteen of 42 infants) was significantly greater than for standard genetic tests (2%, one of 42; P = .0015). Eleven (26%) infants with diagnostic rWGS avoided morbidity, one had a 43% reduction in likelihood of mortality, and one started palliative care. In six of the eleven infants, the changes in management reduced inpatient cost by $800,000-$2,000,000. The findings replicated a prior study of the clinical utility of rWGS in acutely ill inpatient infants, and demonstrated improved outcomes, net healthcare savings and consideration as a first tier test in this setting.
A 2018 review of 36 publications found the cost for whole genome sequencing to range from $1,906USD to $24,810USD and have a wide variance in diagnostic yield from 17% to 73% depending on patient groups.
Rare variant association study
Whole genome sequencing studies enable the assessment of associations between complex traits and both coding and noncoding rare variants ( minor allele frequency (MAF) < 1%) across the genome. Single-variant analyses typically have low power to identify associations with rare variants, and variant set tests have been proposed to jointly test the effects of given sets of multiple rare variants. SNP annotations help to prioritize rare functional variants, and incorporating these annotations can effectively boost the power of genetic association of rare variants analysis of whole genome sequencing studies. Some tools have been specifically developed to provide all-in-one rare variant association analysis for whole-genome sequencing data, including integration of genotype data and their functional annotations, association analysis, result summary and visualization. Meta-analysis of whole genome sequencing studies provides an attractive solution to the problem of collecting large sample sizes for discovering rare variants associated with complex phenotypes. Some methods have been developed to enable functionally informed rare variant association analysis in biobank-scale cohorts using efficient approaches for summary statistic storage.Oncology
In this field, whole genome sequencing represents a great set of improvements and challenges to be faced by the scientific community, as it makes it possible to analyze, quantify and characterize circulating tumor DNA (ctDNA) in the bloodstream. This serves as a basis for early cancer diagnosis, treatment selection and relapse monitoring, as well as for determining the mechanisms of resistance,metastasis
Metastasis is a pathogenic agent's spreading from an initial or primary site to a different or secondary site within the host's body; the term is typically used when referring to metastasis by a cancerous tumor. The newly pathological sites, ...
and phylogenetic
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical dat ...
patterns in the evolution of cancer. It can also help in the selection of individualized treatments for patients suffering from this pathology and observe how existing drugs are working during the progression of treatment. Deep whole genome sequencing involves a subclonal reconstruction based on ctDNA in plasma that allows for complete epigenomic and genomic profiling, showing the expression of circulating tumor DNA in each case.
Newborn screening
In 2013, Green and a team of researchers launched the BabySeq Project to study the ethical and medical consequences of sequencing a newborn's DNA. As of 2015, whole genome and exome sequencing as anewborn screening
Newborn screening (NBS) is a public health program of screening (medicine), screening in infants shortly after birth for conditions that are treatable, but not clinically evident in the newborn period. The goal is to identify infants at risk for ...
tool were deliberated and in 2021, further discussed.
In 2021, the NIH funded BabySeq2, an implementation study that expanded the BabySeq project, enrolling 500 infants from diverse families and track the effects of their genomic sequencing on their pediatric care.
In 2023, the Lancet opined that in the UK "focusing on improving screening by upgrading targeted gene panels might be more sensible in the short term. Whole genome sequencing in the long term deserves thorough examination and universal caution."
Ethical concerns
The introduction of whole genome sequencing may have ethical implications. On one hand, genetic testing can potentially diagnose preventable diseases, both in the individual undergoing genetic testing and in their relatives. On the other hand, genetic testing has potential downsides such as genetic discrimination, loss of anonymity, and psychological impacts such as discovery of non-paternity. Some ethicists insist that the privacy of individuals undergoing genetic testing must be protected, and is of particular concern when minors undergo genetic testing. Illumina's CEO, Jay Flatley, wrongly claimed in February 2009 that "by 2019 it will have become routine to map infants' genes when they are born". This potential use of genome sequencing is highly controversial, as it runs counter to establishedethical
Ethics is the philosophical study of moral phenomena. Also called moral philosophy, it investigates normative questions about what people ought to do or which behavior is morally right. Its main branches include normative ethics, applied e ...
norms for predictive genetic testing
Genetic testing, also known as DNA testing, is used to identify changes in DNA sequence or chromosome structure. Genetic testing can also include measuring the results of genetic changes, such as RNA analysis as an output of gene expression, or ...
of asymptomatic minors that have been well established in the fields of medical genetics
Medical genetics is the branch of medicine that involves the diagnosis and management of hereditary disorders. Medical genetics differs from human genetics in that human genetics is a field of scientific research that may or may not apply to me ...
and genetic counseling. The traditional guidelines for genetic testing have been developed over the course of several decades since it first became possible to test for genetic markers associated with disease, prior to the advent of cost-effective, comprehensive genetic screening.
When an individual undergoes whole genome sequencing, they reveal information about not only their own DNA sequences, but also about probable DNA sequences of their close genetic relatives. This information can further reveal useful predictive information about relatives' present and future health risks. Hence, there are important questions about what obligations, if any, are owed to the family members of the individuals who are undergoing genetic testing. In Western/European society, tested individuals are usually encouraged to share important information on any genetic diagnoses with their close relatives, since the importance of the genetic diagnosis for offspring and other close relatives is usually one of the reasons for seeking a genetic testing in the first place. Nevertheless, a major ethical dilemma can develop when the patients refuse to share information on a diagnosis that is made for serious genetic disorder that is highly preventable and where there is a high risk to relatives carrying the same disease mutation. Under such circumstances, the clinician may suspect that the relatives would rather know of the diagnosis and hence the clinician can face a conflict of interest with respect to patient-doctor confidentiality.
Privacy concerns can also arise when whole genome sequencing is used in scientific research studies. Researchers often need to put information on patient's genotypes and phenotypes into public scientific databases, such as locus specific databases. Although only anonymous patient data are submitted to locus specific databases, patients might still be identifiable by their relatives in the case of finding a rare disease or a rare missense mutation. Public discussion around the introduction of advanced forensic techniques (such as advanced familial searching using public DNA ancestry websites and DNA phenotyping approaches) has been limited, disjointed, and unfocused. As forensic genetics and medical genetics converge toward genome sequencing, issues surrounding genetic data become increasingly connected, and additional legal protections may need to be established.
Public human genome sequences
First people with public genome sequences
The first nearly complete human genomes sequenced were two Americans of predominantlyNorthwestern Europe
Northwestern Europe, or Northwest Europe, is a loosely defined subregion of Europe, overlapping Northern and Western Europe. The term is used in geographic, history, and military contexts.
Geographic definitions
Geographically, Northwestern ...
an ancestry in 2007 ( J. Craig Venter at 7.5-fold coverage, and James Watson
James Dewey Watson (born April 6, 1928) is an American molecular biology, molecular biologist, geneticist, and zoologist. In 1953, he co-authored with Francis Crick the academic paper in ''Nature (journal), Nature'' proposing the Nucleic acid ...
at 7.4-fold). This was followed in 2008 by sequencing of an anonymous Han Chinese
The Han Chinese, alternatively the Han people, are an East Asian people, East Asian ethnic group native to Greater China. With a global population of over 1.4 billion, the Han Chinese are the list of contemporary ethnic groups, world's la ...
man (at 36-fold), a Yoruban man from Nigeria
Nigeria, officially the Federal Republic of Nigeria, is a country in West Africa. It is situated between the Sahel to the north and the Gulf of Guinea in the Atlantic Ocean to the south. It covers an area of . With Demographics of Nigeria, ...
(at 30-fold), a female clinical geneticist ( Marjolein Kriek) from the Netherlands (at 7 to 8-fold), and a female leukemia
Leukemia ( also spelled leukaemia; pronounced ) is a group of blood cancers that usually begin in the bone marrow and produce high numbers of abnormal blood cells. These blood cells are not fully developed and are called ''blasts'' or '' ...
patient in her mid-50s (at 33 and 14-fold coverage for tumor and normal tissues). Steve Jobs
Steven Paul Jobs (February 24, 1955 – October 5, 2011) was an American businessman, inventor, and investor best known for co-founding the technology company Apple Inc. Jobs was also the founder of NeXT and chairman and majority shareholder o ...
was among the first 20 people to have their whole genome sequenced, reportedly for the cost of $100,000. , there were 69 nearly complete human genomes publicly available. In November 2013, a Spanish family made their personal genomics data publicly available under a Creative Commons public domain license. The work was led by Manuel Corpas and the data obtained by direct-to-consumer genetic testing with 23andMe
23andMe Holding Co. is an American personal genomics and biotechnology company based in South San Francisco, California. It is best known for providing a direct-to-consumer genetic testing service in which customers provide a saliva testing, sali ...
and the Beijing Genomics Institute
BGI Group, formerly Beijing Genomics Institute, is a Chinese genomics company with headquarters in Yantian, Shenzhen, Yantian, Shenzhen. The company was originally formed in 1999 as a genetics research center to participate in the Human Genome Pro ...
. This is believed to be the first such Public Genomics dataset for a whole family.
Databases
According to ''Science
Science is a systematic discipline that builds and organises knowledge in the form of testable hypotheses and predictions about the universe. Modern science is typically divided into twoor threemajor branches: the natural sciences, which stu ...
'', the major databases of whole genomes are:
Genomic coverage
In terms of genomic coverage and accuracy, whole genome sequencing can broadly be classified into either of the following: Last updated: November 1, 2021 * A ''draft sequence'', covering approximately 90% of the genome at approximately 99.9% accuracy * A ''finished sequence'', covering more than 95% of the genome at approximately 99.99% accuracy Producing a truly high-quality ''finished'' sequence by this definition is very expensive. Thus, most human "whole genome sequencing" results are ''draft sequences'' (sometimes above and sometimes below the accuracy defined above).See also
* Coverage (genetics) *DNA microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or t ...
* DNA profiling
DNA profiling (also called DNA fingerprinting and genetic fingerprinting) is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is cal ...
* DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
* Duplex sequencing
* Exome Sequencing
Exome sequencing, also known as whole exome sequencing (WES), is a genomic technique for sequencing all of the protein-coding regions of genes in a genome (known as the exome). It consists of two steps: the first step is to select only the subs ...
* Genomics England
* Horizontal correlation
Horizontal correlation is a methodology for gene sequence
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a f ...
* Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
* Lists of sequenced genomes
* Medical genetics
Medical genetics is the branch of medicine that involves the diagnosis and management of hereditary disorders. Medical genetics differs from human genetics in that human genetics is a field of scientific research that may or may not apply to me ...
* Medical Research Council (MRC)
* Nucleic acid sequence
A nucleic acid sequence is a succession of Nucleobase, bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the orde ...
* Personal Genome Project
* Personalized medicine
* Predictive medicine
* Rare functional variant
* Single cell sequencing
* SNP annotation
References
External links
James Watson's Personal Genome Sequence
AAAS/Science: Genome Sequencing Poster
{{Breakthrough of the Year Molecular biology DNA sequencing Biotechnology Genomics Bioinformatics DNA * Gene tests Molecular genetics