Exo-endonuclease on:
[Wikipedia]
[Google]
[Amazon]
In
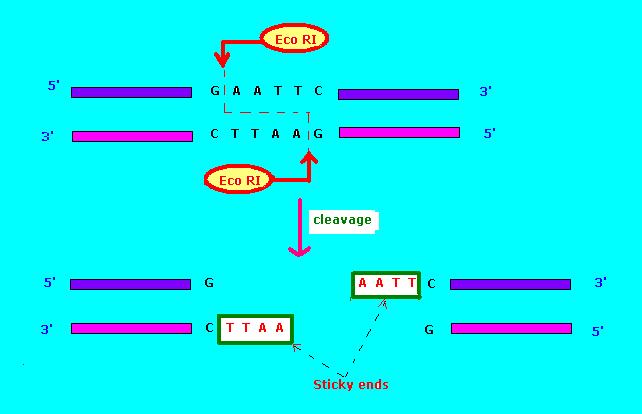 Restriction endonucleases come in several types. A restriction endonuclease typically requires a recognition site and a cleavage pattern (typically of nucleotide bases: A, C, G, T). If the recognition site is outside the region of the cleavage pattern, then the restriction endonuclease is referred to as Type I. If the recognition sequence overlaps with the cleavage sequence, then the restriction endonuclease
Restriction endonucleases come in several types. A restriction endonuclease typically requires a recognition site and a cleavage pattern (typically of nucleotide bases: A, C, G, T). If the recognition site is outside the region of the cleavage pattern, then the restriction endonuclease is referred to as Type I. If the recognition sequence overlaps with the cleavage sequence, then the restriction endonuclease

molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
, endonucleases are enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s that cleave
Cleave may refer to:
* Cleave (surname)
* Cleave (fiber), a controlled break in optical fiber
* RAF Cleave, was an airfield in the north of Cornwall, England, May 1939 - Nov 1945
*The process of protein cleaving as a form of post-translational mod ...
the phosphodiester bond
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is d ...
within a polynucleotide
In molecular biology, a polynucleotide () is a biopolymer composed of nucleotide monomers that are covalently bonded in a chain. DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are examples of polynucleotides with distinct biological func ...
chain (namely DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
or RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
). Some, such as deoxyribonuclease I
Deoxyribonuclease I (usually called DNase I), is an endonuclease of the DNase family coded by the human gene DNASE1.
DNase I is a nuclease that cleaves DNA preferentially at phosphodiester linkages adjacent to a pyrimidine nucleotide, yielding 5 ...
, cut DNA relatively nonspecifically (with regard to sequence), while many, typically called ''restriction endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
'' or ''restriction enzymes'', cleave only at very specific nucleotide sequences. Endonucleases differ from exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
s, which cleave the ends of recognition sequences instead of the middle (''endo'') portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
Restriction enzymes are endonucleases from eubacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the ...
and archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the ''restriction site''. Typically, a restriction site will be a palindromic
A palindrome ( /ˈpæl.ɪn.droʊm/) is a word, number, phrase, or other sequence of symbols that reads the same backwards as forwards, such as ''madam'' or '' racecar'', the date " 02/02/2020" and the sentence: "A man, a plan, a canal – Pana ...
sequence about four to six nucleotides long. Most restriction endonucleases cleave the DNA strand unevenly, leaving complementary single-stranded ends. These ends can reconnect through hybridization and are termed "sticky ends". Once paired, the phosphodiester bonds of the fragments can be joined by DNA ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such ...
. There are hundreds of restriction endonucleases known, each attacking a different restriction site. The DNA fragments cleaved by the same endonuclease can be joined regardless of the origin of the DNA. Such DNA is called recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be fo ...
; DNA formed by the joining of genes into new combinations. ''Restriction endonucleases'' (restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
s) are divided into three categories, Type I, Type II, and Type III, according to their mechanism of action. These enzymes are often used in genetic engineering
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of Genetic engineering techniques, technologies used to change the genet ...
to make recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be fo ...
for introduction into bacterial, plant, or animal cells, as well as in synthetic biology
Synthetic biology (SynBio) is a multidisciplinary field of science that focuses on living systems and organisms. It applies engineering principles to develop new biological parts, devices, and systems or to redesign existing systems found in nat ...
. One of the more famous endonucleases is Cas9
Cas9 (CRISPR associated protein 9, formerly called Cas5, Csn1, or Csx12) is a 160 dalton (unit), kilodalton protein which plays a vital role in the immunological defense of certain bacteria against DNA viruses and plasmids, and is heavily utili ...
.
Categories
Ultimately, there are three categories ofrestriction endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
that relatively contribute to the cleavage of specific sequences. The types I and III are large multisubunit complexes that include both the endonucleases
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while many, t ...
and methylase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Ro ...
activities. Type I can cleave at random sites of about 1000 base pairs or more from the recognition sequence and it requires ATP as source of energy. Type II behaves slightly differently and was first isolated by Hamilton Smith in 1970. They are simpler versions of the endonucleases and require no ATP in their degradation processes. Some examples of type II restriction endonucleases include ''Bam''HI, ''Eco''RI, ''Eco''RV, ''Hin''dIII, and ''Hae''III. Type III, however, cleaves the DNA at about 25 base pairs from the recognition sequence and also requires ATP in the process.
Notations
The commonly used notation for restriction endonucleases is of the form "''Vwx''yZ", where "''Vwx''" are, in italics, the first letter of the genus and the first two letters of the species where this restriction endonuclease may be found, for example, ''Escherichia coli'', ''Eco'', and ''Haemophilus influenzae'', ''Hin''. This is followed by the optional, non-italicized symbol "y", which indicates the type or strain identification, for example, ''Eco''R for ''E. coli'' strains bearing the drug resistance transfer factor RTF-1, ''Eco''B for ''E. coli'' strain B, and ''Hin''d for ''H. influenzae'' strain d. Finally, when a particular type or strain has several different restriction endonucleases, these are identified by Roman numerals, thus, the restriction endonucleases from ''H. influenzae'' strain d are named ''Hin''dI, ''Hin''dII, ''Hin''dIII, etc. Another example: "''Hae''II" and "''Hae''III" refer to bacterium ''Haemophilus aegyptius'' (strain not specified), restriction endonucleases number II and number III, respectively. The restriction enzymes used in molecular biology usually recognize short target sequences of about 4 – 8 base pairs. For instance, the ''Eco''RI enzyme recognizes and cleaves the sequence 5' – GAATTC – 3'.restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
is Type II.
Processes involved with endonucleases
Endonucleases play a role in many aspects of biological life. Below are a couple examples of processes where endonucleases play a crucial role.DNA repair
Endonucleases play a role in DNA repair.AP endonuclease
Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of t ...
, specifically, catalyzes the incision of DNA exclusively at AP sites, and therefore prepares DNA for subsequent excision, repair synthesis and DNA ligation. For example, when depurination occurs, this lesion leaves a deoxyribose sugar with a missing base. The AP endonuclease recognizes this sugar and essentially cuts the DNA at this site and then allows for DNA repair to continue. ''E. coli'' cells contain two AP endonucleases: endonuclease IV (endoIV) and exonuclease III (exoIII) while in eukaryotes, there is only one AP endonuclease.

DNA crosslink repair
Repair of DNA in which the two complementary strands are joined by an interstrand covalent crosslink requires multiple incisions in order to disengage the strands and remove the damage. Incisions are required on both sides of the crosslink and on both strands of the duplex DNA. In mouse embryonic stem cells, an intermediate stage of crosslink repair involves production of double-strand breaks.MUS81
Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the ''MUS81'' gene.
In mammalian somatic cells, MUS81 and another structure specific DNA endonuclease, XPF (ERCC4), play overlapping and essential roles in completio ...
/ EME1 is a structure specific endonuclease involved in converting interstrand crosslinks to double-strand breaks in a DNA replication-dependent manner. After introduction of a double-strand break, further steps are required to complete the repair process. If a crosslink is not properly repaired it can block DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
.
Thymine dimer repair
Exposure of bacteriophage (phage) T4 toultraviolet
Ultraviolet radiation, also known as simply UV, is electromagnetic radiation of wavelengths of 10–400 nanometers, shorter than that of visible light, but longer than X-rays. UV radiation is present in sunlight and constitutes about 10% of ...
irradiation induces thymine dimers
Pyrimidine dimers represent molecular lesions originating from thymine or cytosine bases within DNA, resulting from photochemical reactions. These lesions, commonly linked to direct DNA damage, are induced by ultraviolet light (UV), particularly ...
in the phage DNA. The phage T4 ''denV'' gene encodes endonuclease V
Endonuclease V ''(endoV)'' is a highly conserved endonuclease enzyme family. The primary function of endoV differs significantly in prokaryotes and eukaryotes, as suggested by studies on the ''E. coli'' and human orthologs.
In prokaryotes endoV ...
that catalyzes the initial steps in the repair of these UV-induced thymine dimers. Endonuclease V first cleaves the glycosylic bond on the 5’ side of a pyrimidine dimer and then catalyzes cleavage of the DNA phosphodiester bond that originally linked the two nucleotides of the dimer. Subsequent steps in the repair process involve removal of the dimer remnants and repair synthesis to fill in the resulting single-strand gap using the undamaged strand as template.
Apoptosis
During apoptosis, Apoptotic endonuclease DFF40 is activated to initiate controlled cellular disassembly. This disintegration is characterized by the cleavage of genomic DNA into specific fragments. The precise role of endonucleases in this context is to cleave the DNA at specific sites, generating fragments with defined lengths. These fragments are then packaged into apoptotic bodies, ensuring a neat and efficient removal of the dying cell without causing inflammation or damage to neighboring cells.DNA Replication
Flap endonuclease 1 (FEN1) and Dna2 endonuclease are integral toDNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
on the lagging strand, participating in crucial processes such as primer removal and Okazaki Okazaki may refer to:
*Okazaki (surname)
*Okazaki, Aichi, a city in Japan
*Okazaki Castle, a castle in Japan
*Okazaki fragments
Okazaki fragments are short sequences of DNA nucleotides (approximately 150 to 200 base pairs long in eukaryotes) w ...
fragment processing. Endonucleases are actively involved in processing these fragments by cleaving the phosphodiester bonds between them. This process is integral to the seamless synthesis and joining of Okazaki fragments, contributing to the overall continuity of the newly replicated DNA strand.
RNA Processing
Endonucleases, more specificallyendoribonuclease
In biochemistry, an endoribonuclease is a class of enzyme which is a type of ribonuclease (an RNA cleaver), itself a type of endonuclease (a nucleotide cleaver). It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Ex ...
, play a crucial role in RNA processing, a fundamental step in gene expression. This process involves the precise cleavage of precursor RNA molecules, guided by endonucleases, to generate functional RNAs essential for various cellular functions. Endonucleases selectively cleave precursor RNAs at specific sites, defining the boundaries of functional RNA segments during RNA processing. The outcome of RNA processing is the production of functional RNA molecules, such as transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). Endonucleases contribute to the precision of this process, ensuring the formation of mature and functional RNA species.
Endonucleases like RNase P
Ribonuclease P (, ''RNase P'') is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its functio ...
and tRNase Z (ELAC2), shape precursor tRNAs into mature, functional tRNAs, crucial for accurate translation during protein synthesis. In ribosome biogenesis, endonucleases from the RNase III family, like DROSHA
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the ''DROSHA'' (formerly ''RNASEN'') gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and ...
, play a role in processing precursor rRNAs, contributing to the assembly of functional ribosomes.
DICER
Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into shor ...
and DROSHA
Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the ''DROSHA'' (formerly ''RNASEN'') gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and ...
also from the RNase III family play a role in the processing pre-miRNA to functional miRNA.
Maturation of Nails and Hairs
The endonucleaseDNase1L2
Deoxyribonuclease-1-like 2 is an enzyme that in humans is encoded by the ''DNASE1L2'' gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA th ...
also contribute prominently to the removal of DNA during the formation of hair and nails. This process is essential for the maturation of hair and nail structures and is crucial for the transformation of cells into durable and keratin
Keratin () is one of a family of structural fibrous proteins also known as ''scleroproteins''. It is the key structural material making up Scale (anatomy), scales, hair, Nail (anatomy), nails, feathers, horn (anatomy), horns, claws, Hoof, hoove ...
ized structures, ensuring the strength and integrity of hair and nails.
Further discussion
Restriction endonucleases may be found that cleave standard dsDNA (double-stranded DNA), or ssDNA (single-stranded DNA), or even RNA. This discussion is restricted to dsDNA; however, the discussion can be extended to the following: * Standard dsDNA * Non-standard DNA #Holliday junction
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the j ...
s
# Triple-stranded DNA
Triple-stranded DNA (also known as H-DNA or Triplex-DNA) is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a Nucleic acid double helix#Helix geomet ...
, quadruple-stranded DNA (G-quadruplex
In molecular biology, G-quadruplex secondary structures (G4) are formed in nucleic acids by sequences that are rich in guanine. They are helical in shape and contain guanine tetrads that can form from one, two or four strands. The unimolecular ...
)
# Double-stranded hybrids of DNA and RNA (one strand is DNA, the other strand is RNA)
# Synthetic or artificial DNA (for example, containing bases other than A, C, G, T, refer to the work of Eric T. Kool). Research with synthetic codons
Genetic code is a set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links pro ...
, refer to the research by S. Benner, and enlarging the amino acid set in polypeptides, thus enlarging the proteome or proteomics
Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replicatio ...
, see the research by P. Schultz.
In addition, research is now underway to construct synthetic or artificial restriction endonucleases, especially with recognition sites that are unique within a genome.
Restriction endonucleases or restriction enzymes
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
typically cleave in two ways: blunt-ended or sticky-ended patterns. An example of a Type I restriction endonuclease.
Furthermore, there exist DNA/RNA non-specific endonucleases, such as those that are found in ''Serratia marcescens
''Serratia marcescens'' () is a species of bacillus (shape), rod-shaped, Gram-negative bacteria in the family Yersiniaceae. It is a facultative anaerobe and an opportunistic pathogen in humans. It was discovered in 1819 by Bartolomeo Bizio in Pa ...
'', which act on dsDNA, ssDNA, and RNA.
Common endonucleases
Below are tables of common prokaryotic and eukaryotic endonucleases.Mutations
Xeroderma pigmentosa is a rare, autosomal recessive disease caused by a defective UV-specific endonuclease. Patients with mutations are unable to repair DNA damage caused by sunlight. Sickle Cell anemia is a disease caused by a point mutation. The sequence altered by the mutation eliminates the recognition site for the restriction endonuclease MstII that recognizes the nucleotide sequence. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Pontocerebellar hypoplasias (PCH) represent a group of neurodegenerative autosomal recessive disorders that is caused by mutations in three of the four different subunits of the tRNA-splicing endonuclease complex.See also
*Exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
* Restriction endonuclease
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
* Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and ...
* Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within th ...
* AP endonuclease
Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of t ...
* HUH-tag
HUH endonucleases (HUH-tags) are sequence-specific single-stranded DNA (ssDNA) binding proteins originating from numerous species of bacteria and viruses. Viral HUH endonucleases are involved in initiating rolling circle replication while ones of ...
References
{{Portal bar, Biology, border=no EC 3.1