Elongation Factor Tu on:
[Wikipedia]
[Google]
[Amazon]
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an
 EF-Tu is a
EF-Tu is a
aminoacyl-tRNA
Aminoacyl-tRNA (also aa-tRNA or charged tRNA) is tRNA to which its cognate amino acid is chemically bonded (charged). The aa-tRNA, along with particular elongation factors, deliver the amino acid to the ribosome for incorporation into the polyp ...
(aa-tRNA) to the ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
. It is a G-protein
G proteins, also known as guanine nucleotide-binding proteins, are a family of proteins that act as molecular switches inside cells, and are involved in transmitting signals from a variety of stimuli outside a cell to its interior. Their ...
, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM
Elongation factor Tu, mitochondrial is a protein that in humans is encoded by the ''TUFM'' gene. It is an EF-Tu
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacy ...
.
As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1
eEF-1 are two eukaryotic elongation factors. It forms two complexes, the EF-Tu homolog EF-1A and the EF-Ts homolog EF-1B, the former's guanide exchange factor. Both are also found in archaea.
Structure
The nomenclature for the eEF-1 subuni ...
(EF-1A).
Background
Elongation factors are part of the mechanism that synthesizes newprotein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s through translation in the ribosome. Transfer RNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
s (tRNAs) carry the individual amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s that become integrated into a protein sequence, and have an anticodon
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
for the specific amino acid that they are charged with. Messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA) carries the genetic information that encodes the primary structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
of a protein, and contains codons
Genetic code is a set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links pro ...
that code for each amino acid. The ribosome creates the protein chain by following the mRNA code and integrating the amino acid of an aminoacyl-tRNA (also known as a charged tRNA) to the growing polypeptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ...
chain.
There are three sites on the ribosome for tRNA binding. These are the aminoacyl/acceptor site (abbreviated A), the peptidyl site (abbreviated P), and the exit site (abbreviated E). The P-site holds the tRNA connected to the polypeptide chain being synthesized, and the A-site is the binding site for a charged tRNA with an anticodon complementary to the mRNA codon associated with the site. After binding of a charged tRNA to the A-site, a peptide bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein cha ...
is formed between the growing polypeptide chain on the P-site tRNA and the amino acid of the A-site tRNA, and the entire polypeptide is transferred from the P-site tRNA to the A-site tRNA. Then, in a process catalyzed by the prokaryotic elongation factor EF-G
EF-G (elongation factor G, historically known as translocase) is a prokaryotic elongation factor involved in mRNA translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through th ...
(historically known as translocase), the coordinated translocation of the tRNAs and mRNA occurs, with the P-site tRNA moving to the E-site, where it dissociates from the ribosome, and the A-site tRNA moves to take its place in the P-site.
Biological functions

Protein synthesis
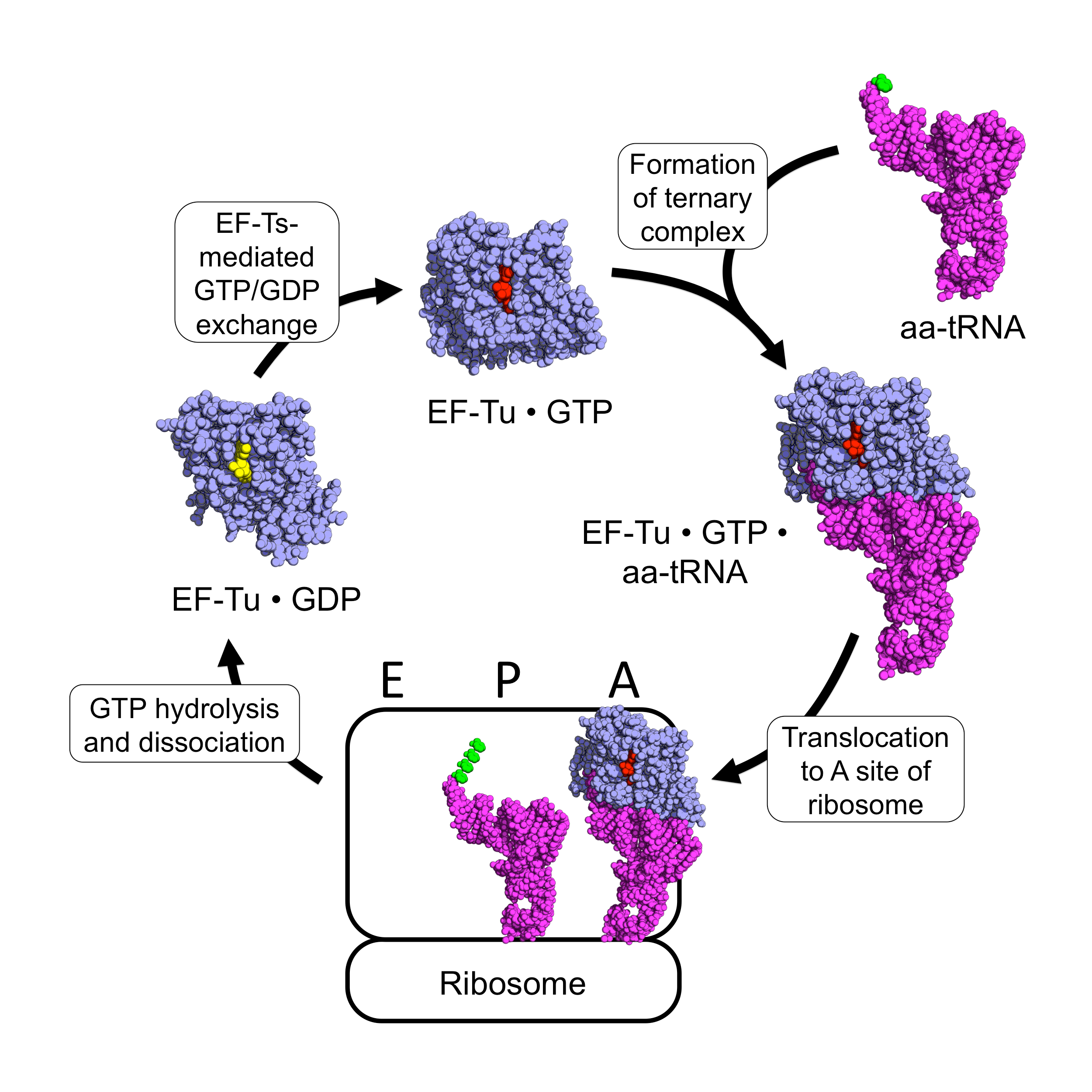
EF-Tu participates in the polypeptide elongation process of protein synthesis. In prokaryotes, the primary function of EF-Tu is to transport the correct aa-tRNA to the A-site of the ribosome. As a G-protein, it uses GTP to facilitate its function. Outside of the ribosome, EF-Tu complexed with GTP (EF-Tu • GTP) complexes with aa-tRNA to form a stable EF-Tu • GTP • aa-tRNAternary complex
A ternary complex is a protein complex containing three different molecules that are bound together. In structural biology, ''ternary complex'' can also be used to describe a crystal containing a protein with two small molecules bound, such as a ...
. EF-Tu • GTP binds all correctly-charged aa-tRNAs with approximately identical affinity, except those charged with initiation residues and selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the ...
. This can be accomplished because although different amino acid residues have varying side-chain properties, the tRNAs associated with those residues have varying structures to compensate for differences in side-chain binding affinities.
The binding of an aa-tRNA to EF-Tu • GTP allows for the ternary complex to be translocated to the A-site of an active ribosome, in which the anticodon of the tRNA binds to the codon of the mRNA. If the correct anticodon binds to the mRNA codon, the ribosome changes configuration and alters the geometry of the GTPase
GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a ...
domain of EF-Tu, resulting in the hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution reaction, substitution, elimination reaction, elimination, and solvation reactions in which water ...
of the GTP associated with the EF-Tu to GDP
Gross domestic product (GDP) is a monetary measure of the total market value of all the final goods and services produced and rendered in a specific time period by a country or countries. GDP is often used to measure the economic performance o ...
and Pi. As such, the ribosome functions as a GTPase-activating protein GTPase-activating proteins or GTPase-accelerating proteins (GAPs) are a family of regulatory proteins whose members can bind to activated G proteins and stimulate their GTPase activity, with the result of terminating the signaling event. GAPs are a ...
(GAP) for EF-Tu. Upon GTP hydrolysis, the conformation of EF-Tu changes drastically and dissociates from the aa-tRNA and ribosome complex. The aa-tRNA then fully enters the A-site, where its amino acid is brought near the P-site's polypeptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ...
and the ribosome catalyzes the covalent transfer of the polypeptide onto the amino acid.
In the cytoplasm, the deactivated EF-Tu • GDP is acted on by the prokaryotic elongation factor EF-Ts
EF-Ts (elongation factor thermo stable) is one of the prokaryotic elongation factors. It is found in human mitochondria as TSFM. It is similar to eukaryotic eEF-1, EF-1B.
EF-Ts serves as the guanine nucleotide exchange factor for EF-Tu (elonga ...
, which causes EF-Tu to release its bound GDP. Upon dissociation of EF-Ts, EF-Tu is able to complex with a GTP due to the 5– to 10–fold higher concentration of GTP than GDP in the cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
, resulting in reactivated EF-Tu • GTP, which can then associate with another aa-tRNA.
Maintaining translational accuracy
EF-Tu contributes to translational accuracy in three ways. In translation, a fundamental problem is that near-cognate anticodons have similar binding affinity to a codon as cognate anticodons, such that anticodon-codon binding in the ribosome alone is not sufficient to maintain high translational fidelity. This is addressed by the ribosome not activating the GTPase activity of EF-Tu if the tRNA in the ribosome's A-site does not match the mRNA codon, thus preferentially increasing the likelihood for the incorrect tRNA to leave the ribosome. Additionally, regardless of tRNA matching, EF-Tu also induces a delay after freeing itself from the aa-tRNA, before the aa-tRNA fully enters the A-site (a process called accommodation). This delay period is a second opportunity for incorrectly charged aa-tRNAs to move out of the A-site before the incorrect amino acid is irreversibly added to the polypeptide chain. A third mechanism is the less well understood function of EF-Tu to crudely check aa-tRNA associations and reject complexes where the amino acid is not bound to the correct tRNA coding for it.Other functions
EF-Tu has been found in large quantities in thecytoskeleton
The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is compos ...
s of bacteria, co-localizing underneath the cell membrane
The cell membrane (also known as the plasma membrane or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of a cell from the outside environment (the extr ...
with MreB
MreB is a protein found in bacteria that has been identified as a homologue of actin, as indicated by similarities in tertiary structure and conservation of active site peptide sequence. The conservation of protein structure suggests the commo ...
, a cytoskeletal element that maintains cell shape. Defects in EF-Tu have been shown to result in defects in bacterial morphology. Additionally, EF-Tu has displayed some chaperone-like characteristics, with some experimental evidence suggesting that it promotes the refolding of a number of denatured proteins ''in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
.'' EF-Tu has been found to moonlight
Moonlight consists of mostly sunlight (with little earthlight) reflected from the parts of the Moon's surface where the Sun's light strikes.
History
The ancient Greek philosopher Anaxagoras was aware that "''the sun provides the moon with its ...
on the cell surface of the pathogenic bacteria ''Staphylococcus aureus
''Staphylococcus aureus'' is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often posi ...
'', ''Mycoplasma pneumoniae
''Mycoplasma pneumoniae'' is a species of very small-cell bacteria that lack a cell wall, in the class Mollicutes. ''M. pneumoniae'' is a human pathogen that causes the disease Mycoplasma pneumonia, a form of atypical bacterial pneumonia related ...
'', and '' Mycoplasma hyopneumoniae'', where EF-Tu is processed and can bind to a range of host molecules. In ''Bacillus cereus
''Bacillus cereus'' is a Gram-positive bacteria, Gram-positive Bacillus, rod-shaped bacterium commonly found in soil, food, and marine sponges. The specific name, ''cereus'', meaning "waxy" in Latin, refers to the appearance of colonies grown o ...
'', EF-Tu also moonlights on the surface, where it acts as an environmental sensor and binds to substance P
Substance P (SP) is an undecapeptide (a peptide composed of a chain of 11 amino acid residues) and a type of neuropeptide, belonging to the tachykinin family of neuropeptides. It acts as a neurotransmitter and a neuromodulator. Substance P ...
.
Structure
 EF-Tu is a
EF-Tu is a monomer
A monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or two- or three-dimensional network in a process called polymerization.
Classification
Chemis ...
ic protein with molecular weight
A molecule is a group of two or more atoms that are held together by Force, attractive forces known as chemical bonds; depending on context, the term may or may not include ions that satisfy this criterion. In quantum physics, organic chemi ...
around 43 kDa
The dalton or unified atomic mass unit (symbols: Da or u, respectively) is a unit of mass defined as of the mass of an unbound neutral atom of carbon-12 in its nuclear and electronic ground state and at rest. It is a non-SI unit accepted f ...
in ''Escherichia coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly fo ...
''. The protein consists of three structural domains: a GTP-binding domain and two oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
-binding domains, often referred to as domain 2 and domain 3. The N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amin ...
domain I of EF-Tu is the GTP-binding domain. It consists of a six beta-strand
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a gener ...
core flanked by six alpha-helices
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix).
The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of l ...
. Domains II and III of EF-Tu, the oligonucleotide-binding domains, both adopt beta-barrel
In protein structures, a beta barrel (β barrel) is a beta sheet (β sheet) composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands ...
structures.
The GTP-binding domain I undergoes a dramatic conformational change upon GTP hydrolysis to GDP, allowing EF-Tu to dissociate from aa-tRNA and leave the ribosome. Reactivation of EF-Tu is achieved by GTP binding in the cytoplasm, which leads to a significant conformational change that reactivates the tRNA-binding site of EF-Tu. In particular, GTP binding to EF-Tu results in a ~90° rotation of domain I relative to domains II and III, exposing the residues of the tRNA-binding active site.
Domain 2 adopts a beta-barrel
In protein structures, a beta barrel (β barrel) is a beta sheet (β sheet) composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands ...
structure, and is involved in binding to charged tRNA. This domain is structurally
A structure is an arrangement and organization of interrelated elements in a material object or system, or the object or system so organized. Material structures include man-made objects such as buildings and machines and natural objects such as ...
related to the C-terminal domain of EF2
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation ...
, to which it displays weak sequence similarity. This domain is also found in other protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s such as translation initiation factor IF-2
Bacterial initiation factor-2 is a bacterial initiation factor.
IF2 binds to an initiator tRNA and controls the entry of tRNA onto the ribosome. IF2, bound to GTP, binds to the 30S P site. After associating with the 30S subunit, fMet-tRNAf bi ...
and tetracycline
Tetracycline, sold under various brand names, is an antibiotic in the tetracyclines family of medications, used to treat a number of infections, including acne, cholera, brucellosis, plague, malaria, and syphilis. It is available in oral an ...
-resistance proteins. Domain 3 represents the C-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When t ...
domain, which adopts a beta-barrel
In protein structures, a beta barrel (β barrel) is a beta sheet (β sheet) composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands ...
structure, and is involved in binding to both charged tRNA and to EF1B (or EF-Ts).
Evolution
The GTP-binding domain is conserved in both EF-1alpha/EF-Tu and also in EF-2/EF-G
EF-G (elongation factor G, historically known as translocase) is a prokaryotic elongation factor involved in mRNA translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through th ...
and thus seems typical for GTP-dependent proteins which bind non-initiator tRNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
s to the ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
. The GTP-binding translation factor
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
family also includes the eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
peptide chain release factor
A release factor is a protein that allows for the termination of Translation (biology), translation by recognizing the termination codon or stop codon in an mRNA sequence. They are named so because they release new peptides from the ribosome.
...
GTP-binding subunits and prokaryotic
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
peptide chain release factor 3 (RF-3); the prokaryotic
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
GTP-binding protein lepA and its homologue in yeast (GUF1) and ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a Hybrid word, blend of the Greek ''caeno-'' (recent), ''r ...
'' (ZK1236.1); yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom (biology), kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are est ...
HBS1; rat
Rats are various medium-sized, long-tailed rodents. Species of rats are found throughout the order Rodentia, but stereotypical rats are found in the genus ''Rattus''. Other rat genera include '' Neotoma'' (pack rats), '' Bandicota'' (bandicoo ...
Eef1a1 (formerly "statin S1"); and the prokaryotic
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
selenocysteine
Selenocysteine (symbol Sec or U, in older publications also as Se-Cys) is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the ...
-specific elongation factor
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation ...
selB.
Disease relevance
Along with the ribosome, EF-Tu is one of the most important targets forantibiotic
An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting pathogenic bacteria, bacterial infections, and antibiotic medications are widely used in the therapy ...
-mediated inhibition of translation. Antibiotics targeting EF-Tu can be categorized into one of two groups, depending on the mechanism of action, and one of four structural families. The first group includes the antibiotics pulvomycin and GE2270A, and inhibits the formation of the ternary complex. The second group includes the antibiotics kirromycin and enacyloxin, and prevents the release of EF-Tu from the ribosome after GTP hydrolysis.
See also
*Prokaryotic elongation factors
Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation ...
* EF-Ts
EF-Ts (elongation factor thermo stable) is one of the prokaryotic elongation factors. It is found in human mitochondria as TSFM. It is similar to eukaryotic eEF-1, EF-1B.
EF-Ts serves as the guanine nucleotide exchange factor for EF-Tu (elonga ...
(elongation factor thermo stable)
* EF-G
EF-G (elongation factor G, historically known as translocase) is a prokaryotic elongation factor involved in mRNA translation. As a GTPase, EF-G catalyzes the movement (translocation) of transfer RNA (tRNA) and messenger RNA (mRNA) through th ...
(elongation factor G)
* EF-P (elongation factor P)
* eEF-1
eEF-1 are two eukaryotic elongation factors. It forms two complexes, the EF-Tu homolog EF-1A and the EF-Ts homolog EF-1B, the former's guanide exchange factor. Both are also found in archaea.
Structure
The nomenclature for the eEF-1 subuni ...
* EFR (EF-Tu receptor) EFR may refer to: Organisations
* Eastern Frontier Rifles, a State Armed Police Force for the Indian state of West Bengal
* École Française de Radioélectricité, now EFREI, a French private engineering school
* Ecological fiscal reform
* Econ ...
References
External links
* * {{GTPases Protein biosynthesis Protein domains