In
molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
, circular ribonucleic acid (or circRNA) is a type of single-stranded
RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
which, unlike linear RNA, forms a
covalently closed continuous loop. In circular RNA, the
3' and 5' ends normally present in an RNA molecule have been joined together. This feature confers numerous properties to circular RNA, many of which have only recently been identified.
Many types of circular RNA arise from otherwise
protein-coding genes. Some circular RNA have been shown to code for
proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, re ...
. Some types of circular RNA have also recently shown potential as
gene regulators. The biological function of most circular RNA is unclear.
Because circular RNA do not have 5' or 3' ends, they are resistant to
exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
-mediated
degradation and are presumably more stable than most linear RNA in cells.
Circular RNA has been linked to some diseases such as
cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
.
RNA splicing
In contrast to genes in
bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
,
eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
genes are split by non-coding sequences called
intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
s. In eukaryotes, as a gene is transcribed from DNA into a
messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
(mRNA) transcript, intervening introns are removed, leaving only
exons
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence i ...
in the mature mRNA, which can subsequently be translated to produce the protein product.
The
spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to sp ...
,
a protein-RNA complex located in the nucleus, catalyzes splicing in the following manner:
# The
spliceosome
A spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs ( snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to sp ...
recognizes an
intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
, which is flanked by specific sequences at its 5' and 3' ends, known as a donor splice site (or 5' splice site) and an acceptor splice site (or 3' splice site), respectively.
# The 5' splice site sequence is then subjected to a
nucleophilic attack by a downstream sequence called the branch point, resulting in a circular structure called a lariat.
# The free 5' exon then attacks the 3' splice site, joining the two exons and releasing a structure known as an ''intron lariat''. The intron lariat is subsequently de-branched and quickly degraded.

Alternative splicing
Alternative splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene ma ...
is a phenomenon through which one RNA transcript can yield different protein products based on which segments are considered "introns" and "exons" during a splicing event.
Although not specific to humans, it is a partial explanation for the fact that humans and other much simpler species (such as nematodes) have similar numbers of genes (in the range of 20 - 25 thousand). One of the most striking examples of alternative splicing is in the ''Drosophila''
DSCAM gene, which can give rise to approximately 30 thousand distinct alternatively spliced isoforms.
Non-canonical splicing
Exon scrambling
Exon scrambling, also called exon shuffling, describes an event in which exons are spliced in a "non-canonical" (atypical) order. There are three ways in which exon scrambling can occur:
#
Tandem exon duplication in the genome, which often occurs in cancers.
#
Trans-splicing
''Trans''-splicing is a special form of RNA processing where exons from two different primary RNA transcripts are joined end to end and ligated. It is usually found in eukaryotes and mediated by the spliceosome, although some bacteria and archa ...
, in which two RNA transcripts fuse, resulting in a linear transcript containing exons that, for example, may be derived from genes encoded on two different chromosomes. Trans-splicing is very common in ''C. elegans''
# A splice donor site being joined to a splice acceptor site further upstream in the primary transcript, yielding a circular transcript.
The notion that circularized transcripts are byproducts from imperfect splicing is supported by the low abundance and the lack of sequence conservation of most circRNAs,
but has been challenged.
[
]
Alu elements impact circRNA splicing
Repetitive Alu sequences represent approximately 10% of the human genome. The presence of Alu elements in flanking introns of protein-coding genes adjacent to the first and last exons that form circRNAs, influence the formation of circRNAs.[ It is important that the flanking intronic Alu elements are complementary, as this enables RNA pairing, which in turn facilitates circRNA synthesis.]
Impact of RNA editing on circRNA formation
RNAs can undergo base modification by RNA editing after transcription. RNA editing occurs mainly in Alu elements of protein-coding genes.
Characteristics of circular RNA
Early discoveries of circRNAs
Early discoveries of circular RNAs led to the belief that they lacked significance due to their rarity. These early discoveries included the analysis of genes like the DCC DCC may refer to:
Biology
* Netrin receptor DCC, human receptor protein, and the gene encoding it
* Dosage compensation complex
Business
* Day Chocolate Company
* DCC plc, an Irish holding company
* Doppelmayr Cable Car, cable car company
* D ...
and Sry genes, and the recent discovery of the human non-coding RNA ANRIL, all of which expressed circular isoforms. CircRNA producing genes like the human ETS-1 gene, the human and rat cytochrome P450 genes, the rat androgen binding protein gene ( Shbg), and the human dystrophin gene were also discovered.
Genome-wide identification of circRNAs
Scrambled isoforms and circRNAs
In 2012, in an effort to initially identify cancer-specific exon scrambling events, scrambled exons were discovered in large numbers in both normal and cancer cells. It was found that scrambled exon isoforms comprised about 10% of the total transcript isoforms in leukocytes
White blood cells (scientific name leukocytes), also called immune cells or immunocytes, are cells of the immune system that are involved in protecting the body against both infectious disease and foreign entities. White blood cells are genera ...
, with 2,748 scrambled isoforms in HeLa
HeLa () is an immortalized cell line used in scientific research. It is the oldest human cell line and one of the most commonly used. HeLa cells are durable and prolific, allowing for extensive applications in scientific study. The line is ...
and H9 embryonic stem cell
Embryonic stem cells (ESCs) are Cell potency#Pluripotency, pluripotent stem cells derived from the inner cell mass of a blastocyst, an early-stage pre-Implantation (human embryo), implantation embryo. Human embryos reach the blastocyst stage 4� ...
s being identified. Additionally, about 1 in 50 expressed genes produced scrambled transcript isoforms at least 10% of the time. Tests used to recognize circularity included treating samples with RNase R, an enzyme that degrades linear but not circular RNAs, and testing for the presence of poly-A tails, which are not present in circular molecules. Overall, 98% of scrambled isoforms were found to represent circRNAs, circRNAs were found to be located in the cytoplasm, and circRNAs were found to be abundant.
Discovery of a higher abundance of circRNAs
In 2013, a higher abundance of circRNAs was discovered. Human fibroblast
A fibroblast is a type of cell (biology), biological cell typically with a spindle shape that synthesizes the extracellular matrix and collagen, produces the structural framework (Stroma (tissue), stroma) for animal Tissue (biology), tissues, and ...
RNA was treated with RNase R to enrich for circular RNAs, followed by the categorization of circular transcripts based on their abundance (low, medium, high).[ Approximately 1 in 8 expressed genes were found to produce detectable levels of circRNAs, including those of low abundance, which was significantly higher than previously suspected, and was attributed to greater sequencing depth.]
CircRNAs tissue specificity and antagonist activity
At the same time, a computational method to detect circRNAs was developed, leading to de novo detection of circRNAs in humans, mice, and ''C. elegans'', and extensively validating them. The expression of circRNAs was often found to be tissue/developmental stage specific. Additionally, circRNAs were found to have the ability to act as antagonists of miRNAs, microRNAs which interfere with translation of mRNAs, as exemplified by the circRNA CDR1as, which has miRNA binding sites (as seen below).
CircRNAs and ENCODE Ribozero RNA-seq data
In 2014, human circRNAs were identified and quantified from ENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims "to build a comprehensive parts list of functional elements in the human genome."
ENCODE also supports further biomedical research by "generating community resourc ...
Ribozero RNA-seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
data. Most circRNAs were found to be minor splice isoforms and to be expressed in only a few cell types, with 7,112 human circRNAs having circular fractions (the fraction of similarity an isoform has to transcripts the same locus) of at least 10%. CircRNAs were also found to be no more conserved than their linear controls and, according to ribosome profiling, are not translated.<
CircRNAs and CIRCexplorer
In the same year, CIRCexplorer, a tool used to identify thousands of circRNAs in humans without RNase R RNA-seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also k ...
data, was developed. The vast majority of identified highly expressed exonic circular RNAs were found to be processed from exons located in the middle of RefSeq genes, suggesting that the circular RNA formation is generally coupled to RNA splicing. It was determined that most circular RNAs contain multiple, most commonly, two to three, exons. Exons from circRNAs with only one circularized exon were found to be much longer than those from circRNAs with multiple circularized exons, indicating that processing may prefer a certain length to maximize exon(s) circularization. The introns of circularized exons generally contain high Alu densities that can form inverted repeated Alu pairs (IRAlus). IRAlus, either convergent or divergent, are juxtaposed across flanking introns of circRNAs in a parallel way with similar distances to adjacent exons. IRAlus, and other non-repetitive, but complementary, sequences were also found to promote circular RNA formation. On the other hand, exon circularization efficiency was determined to be affected by the competition of RNA pairing, such that alternative RNA pairing, and its competition, leads to alternative circularization. Finally, both exon circularization and its regulation were found to be evolutionarily dynamic.[
]
Genome-wide calling of circRNA in Alzheimer disease cases
The Cruchaga lab performed the first large scale analyses of circRNA in Alzheimer disease (AD) and demonstrated the role of circRNAs in health and disease. A total of 148 circRNAs were found to be significantly associated in multiple datasets with Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disease and the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As the disease advances, symptoms can include problems wit ...
status and clinical dementia rating (CDR) at death after false discovery rate
In statistics, the false discovery rate (FDR) is a method of conceptualizing the rate of type I errors in null hypothesis testing when conducting multiple comparisons. FDR-controlling procedures are designed to control the FDR, which is the exp ...
(FDR) correction. The expression of circRNAs was independent of the lineal form and that circRNA expression was also corrected by cell proportion. CircRNAs were also found to be co-expressed with known causal Alzheimer genes, such as APP and PSEN1
Presenilin-1 (PS-1) is a presenilin protein that in humans is encoded by the ''PSEN1'' gene. Presenilin-1 is one of the four core proteins in the gamma secretase complex, which is considered to play an important role in generation of amyloid bet ...
, indicating that some circRNAs are also part of the causal pathway. Altogether, circRNA brain expression was found to explain more about Alzheimer's clinical manifestations than the number of APOε4 alleles, suggesting that circRNAs could be used as a potential biomarker for Alzheimer's.
Classes of CircRNA
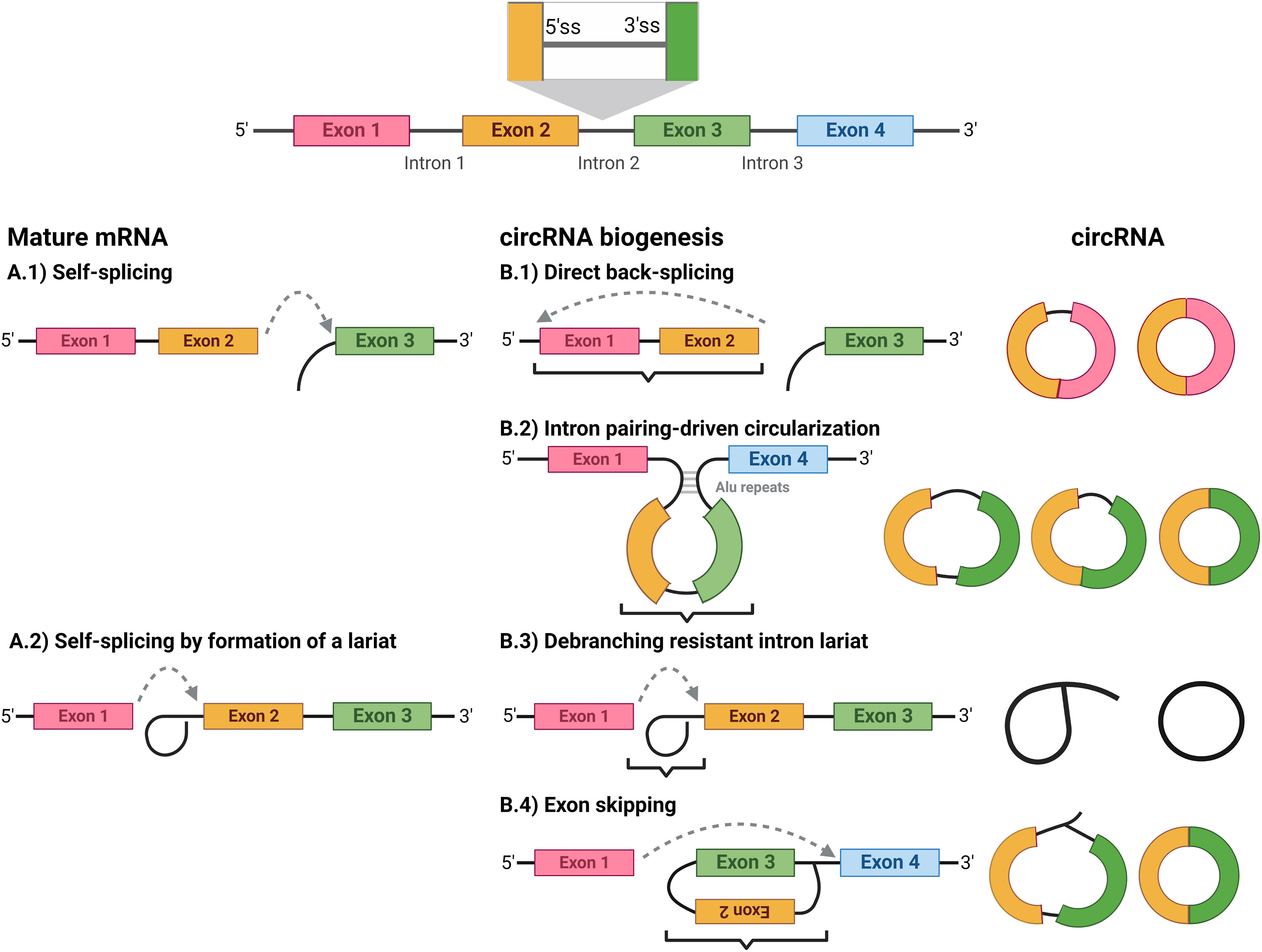
 Circular RNAs can be separated into five classes:
Circular RNAs can be separated into five classes:
Length of circRNAs
A recent study of human circRNAs revealed that these molecules are usually composed of 1–5 exons.protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
,
Location of circRNAs in the cell
In the cell, circRNAs are predominantly found in the cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
, where the number of circular RNA transcripts derived from a gene can be up to ten times greater than the number of associated linear RNAs generated from that locus. It is unclear how circular RNAs exit the nucleus through a relatively small nuclear pore
The nuclear pore complex (NPC), is a large protein complex giving rise to the nuclear pore. A great number of nuclear pores are studded throughout the nuclear envelope that surrounds the eukaryote cell nucleus. The pores enable the nuclear tran ...
. Because the nuclear envelope
The nuclear envelope, also known as the nuclear membrane, is made up of two lipid bilayer membranes that in eukaryotic cells surround the nucleus, which encloses the genetic material.
The nuclear envelope consists of two lipid bilayer membran ...
breaks down during mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
, one hypothesis is that the molecules exit the nucleus during this phase of the cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
.
CircRNAs are more stable than linear RNAs
CircRNAs lack a polyadenylated tail and, therefore, are predicted to be less prone to degradation by exonucleases. In 2015, Enuka ''et al.'' measured the half-lives of 60 circRNAs and their linear counterparts expressed from the same host gene and revealed that the median half-life of circRNAs of mammary cells (18.8 to 23.7 hours) is at least 2.5 times longer than the median half-life of their linear counterparts (4.0 to 7.4 hours).
Plausible functions of circular RNA
Evolutionary conservation of circularization mechanisms and signals
CircRNAs have been identified in various species across the domains of life. In 2011, Danan ''et al.'' sequenced RNA from Archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
. After digesting total RNA with RNase R, they were able to identify circular species, indicating that circRNAs are not specific to eukaryotes. However, these archaeal circular species are probably not made via splicing, suggesting that other mechanisms to generate circular RNA likely exist.
CircRNAs were found to be largely conserved between human and sheep. By analyzing total RNA sequencing data from sheep's parietal lobe cortex and peripheral blood mononuclear cells it was shown that 63% of the detected circRNAs are homologous to known human circRNAs.
 In a closer evolutionary connection, a comparison of RNA from mouse testes vs. RNA from a human cell found 69 orthologous circRNAs. For example, both humans and mice encode the HIPK2 and HIPK3 genes, two paralogous kinases which produce a large amount of circRNA from one particular exon in both species.
In a closer evolutionary connection, a comparison of RNA from mouse testes vs. RNA from a human cell found 69 orthologous circRNAs. For example, both humans and mice encode the HIPK2 and HIPK3 genes, two paralogous kinases which produce a large amount of circRNA from one particular exon in both species.
CDR1as/CiRS-7 as a miR-7 sponge
microRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcr ...
s (miRNAs) are small (~21nt) non-coding RNAs that repress translation of messenger RNAs involved in a large, diverse set of biological processes. They directly base-pair to target messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
s (mRNAs), and can trigger cleavage of the mRNA depending on the degree of complementarity.
MicroRNAs are grouped in "seed families". Family members share nucleotides 2–7, known as the seed region.antisense
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, ...
to the human CDR1 (gene) locus (hence the name CDR1as),zebrafish
The zebrafish (''Danio rerio'') is a species of freshwater ray-finned fish belonging to the family Danionidae of the order Cypriniformes. Native to South Asia, it is a popular aquarium fish, frequently sold under the trade name zebra danio (an ...
, which do not have the CDR1 locus in their genome, provides evidence for CiRS-7's sponge activity. During development, miR-7 is strongly expressed in the zebrafish brain. To silence miR-7 expression in zebrafish, Memczak and colleagues took advantage of a tool called morpholino
A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule (colloquially, an oligo) used in molecular biology to modify gene expression. Its molecular structure contains ...
, which can base pair and sequester target molecules. Morpholino treatment had the same severe effect on midbrain development as ectopically expressing CiRS-7 in zebrafish brains using injected plasmids
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
. This indicates a significant interaction between CiRS-7 and miR-7 in vivo.inverted repeat
An inverted repeat (or IR) is a single stranded sequence of nucleotides followed downstream by its complementarity (molecular biology), reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complem ...
s (IRs) over 15.5 kilobases (kb) in length. When one or both of the IRs are deleted, circularization does not occur. It was this finding that introduced the idea of inverted repeats enabling circularization.
Because circular RNA sponges are characterized by high expression levels, stability, and a large number of miRNA binding sites, they are likely to be more effective sponges than those that are linear.
Other possible functions for circRNAs
Though recent attention has been focused on circRNA's "sponge" functions, scientists are considering several other functional possibilities as well. For example, some areas of the mouse adult hippocampus
The hippocampus (: hippocampi; via Latin from Ancient Greek, Greek , 'seahorse'), also hippocampus proper, is a major component of the brain of humans and many other vertebrates. In the human brain the hippocampus, the dentate gyrus, and the ...
show expression of CiRS-7 but not miR-7, suggesting that CiRS-7 may have roles that are independent of interacting with the miRNA.RNA-binding protein
RNA-binding proteins (often abbreviated as RBPs) are proteins that bind to the double or single stranded RNA in cell (biology), cells and participate in forming ribonucleoprotein complexes.
RBPs contain various structural motifs, such as RNA reco ...
s (RBPs) and RNAs besides miRNAs to form RNA-protein complexes.
Circular intronic long non-coding RNAs (ciRNAs)
Usually, intronic lariats (see above) are debranched and rapidly degraded. However, a debranching failure can lead to the formation of circular intronic long non-coding RNAs, also known as ciRNAs.cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
. In addition, these molecules contain few (if any) miRNA binding sites. Instead of acting as sponges, ciRNAs seem to function in regulating the expression of their parent genes. For example, a relatively abundant ciRNA called ci-ankrd52 positively regulates Pol II transcription. Many ciRNAs remain at their "sites of synthesis" in the nucleus. However, ciRNA may have roles other than simply regulating their parent genes, as ciRNAs do localize to additional sites in the nucleus other than their "sites of synthesis".
Circular RNA and disease
As with most topics in molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
, it is important to consider how circular RNA can be used as a tool to help mankind. Given its abundance, evolutionary conservation, and potential regulatory role, it is worthwhile to look into how circular RNA can be used to study pathogenesis
In pathology, pathogenesis is the process by which a disease or disorder develops. It can include factors which contribute not only to the onset of the disease or disorder, but also to its progression and maintenance. The word comes .
Descript ...
and devise therapeutic interventions. For example:
* Circular ANRIL (cANRIL) is the circular form of ANRIL, a long non-coding RNA (ncRNA). Expression of cANRIL is correlated with risk for atherosclerosis
Atherosclerosis is a pattern of the disease arteriosclerosis, characterized by development of abnormalities called lesions in walls of arteries. This is a chronic inflammatory disease involving many different cell types and is driven by eleva ...
, a disease in which the arteries become hard. It has been proposed that cANRIL can modify INK4/ARF expression, which, in turn, increases risk for atherosclerosis. Further study of cANRIL expression could potentially be used to prevent or treat atherosclerosis.
* miR-7 plays an important regulatory role in several cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
s and in Parkinson's disease
Parkinson's disease (PD), or simply Parkinson's, is a neurodegenerative disease primarily of the central nervous system, affecting both motor system, motor and non-motor systems. Symptoms typically develop gradually and non-motor issues become ...
, which is a brain illness of unknown origin.transgene
A transgene is a gene that has been transferred naturally, or by any of a number of genetic engineering techniques, from one organism to another. The introduction of a transgene, in a process known as transgenesis, has the potential to change the ...
, which is a synthetic gene that is transferred between organisms. It is also important to consider how transgenes can be expressed only in specific tissues, or expressed only when induced.
Circular RNAs play a role in Alzheimer disease pathogenesis
Dube ''et al''.,[ demonstrated for the first time that brain circular RNAs (circRNA) are part of the pathogenic events that lead to ]Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disease and the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As the disease advances, symptoms can include problems wit ...
, hypothesizing that specific circRNA would be differentially expressed in AD cases compared to controls and that those effects could be detected early in the disease. They optimized and validated a novel analyses pipeline for circular RNAs (circRNA). They performed a three-stage study design, using the Knight ADRC brain RNA-seq data as discovery (stage 1), using the data from Mount Sinai as replication (stage 2) and a meta-analysis (stage 3) to identify the most significant circRNA differentially expressed in Alzheimer disease. Using his pipeline, they found 3,547 circRNA that passed stringent QC in the Knight ADRC cohort that includes RNA-seq from 13 controls and 83 Alzheimer cases, and 3,924 circRNA passed stringent QC in the MSBB dataset. A meta-analysis of the discovery and replication results revealed a total of 148 circRNAs that were significantly correlated with CDR after FDR correction. In addition, 33 circRNA passed the stringent gene-based, Bonferroni multiple test correction of 5×10-6, including circHOMER1 (P =2.21×10−18) and circCDR1-AS (P = 2.83 × 10−8), among others. They also performed additional analyses to demonstrate that the expression of circRNA were independent of the lineal form as well as the cell proportion that can confound the brain RNA-seq analyses in Alzheimer disease studies. They performed co-expression analyses of all the circRNA together with the lineal forms and found that circRNA, including those that were differentially expressed in Alzheimer disease compared to controls co-expressed with known causal Alzheimer genes, such as APP and PSEN1, indicating that some circRNA are also part of the causal pathway. They also demonstrated that circRNA brain expression explained more about Alzheimer clinical manifestations that the number of APOε4 alleles, suggesting that could be used as a potential biomarker for Alzheimer disease. This is an important study for the field, as it is the first time that circRNA are quantified and validated (by real-time PCR) in human brain samples at genome-wide scale and in large and well-characterized cohorts. It also demonstrates that these RNA forms are likely to be implicated on complex traits including Alzheimer disease will help to understand the biological events that leads to disease.
Circular RNA pathogenesis in Heart disease, Renal disease, Liver disease, autoimmune disease, and Cancer development
Heart disease
Recent studies have shown that circRNA is associated with heart failure and heart disease. circFOXO3, Titin genes, circSLC8A1-1 and circAmotl1 play an important role in cardiac function through upregulation or inhibition relevant to heart disease. Overexpression of circFOXO3 and its downregulation binds to the transcription factors E2F1, HIF1α and protein ID1, FAK, causing cardiomyopathy induced by DOX. Titin gene derived circRNA induces cardiotoxicity in cardiomyocytes. circSLC8A1-1 overexpression causes sponging of the cardiac hypertrophy regulator miR-133 and leads to heart failure. Apart from circRNA-mediated cardiac disease, some circRNAs have played a role in cardiac damage repair. For example, circAmotl1 overexpression increases cardiomyocyte longevity through binding and translocation of AKT that regulates cardiac repair. Circular RNA CDR1 has an important role during infection in the myocardium
Cardiac muscle (also called heart muscle or myocardium) is one of three types of vertebrate muscle tissues, the others being skeletal muscle and smooth muscle. It is an involuntary, striated muscle that constitutes the main tissue of the wall o ...
. Cardiac dysfunction occurs post myocardial infection due to CircNfix downregulation. Since various types of circular RNA are related to heart disease, it can be used as a potential biomarker
In biomedical contexts, a biomarker, or biological marker, is a measurable indicator of some biological state or condition. Biomarkers are often measured and evaluated using blood, urine, or soft tissues to examine normal biological processes, ...
and therapeutic target. For example, postoperative atrial fibrillation
Atrial fibrillation (AF, AFib or A-fib) is an Heart arrhythmia, abnormal heart rhythm (arrhythmia) characterized by fibrillation, rapid and irregular beating of the Atrium (heart), atrial chambers of the heart. It often begins as short periods ...
has been observed in some patients after cardiac surgery where circRNA_025016 is used as a biomarker. Although the relevance of circular RNA overexpression and downregulation to heart disease has been found from various research studies, it is still unclear. Therefore, further research is needed to trace disease progression in different stages of cardiac dysfunction using circular RNA as a biomarker and can be used for gene delivery purposes in cells.
Renal disease
Various studies have demonstrated that circular RNA acts as a prognostic agent and biomarker in kidney diseases including renal cell carcinoma
Carcinoma is a malignancy that develops from epithelial cells. Specifically, a carcinoma is a cancer that begins in a tissue that lines the inner or outer surfaces of the body, and that arises from cells originating in the endodermal, mesoder ...
, acute kidney injury, diabetic nephropathy
Diabetic nephropathy, also known as diabetic kidney disease, is the chronic loss of kidney function occurring in those with diabetes mellitus. Diabetic nephropathy is the leading cause of chronic kidney disease (CKD) and end-stage renal disease ...
, and lupus nephritis
Lupus nephritis is an inflammation of the kidneys caused by systemic lupus erythematosus (SLE) and childhood-onset systemic lupus erythematosus which is a more severe form of SLE that develops in children up to 18 years old; both are autoimmune d ...
. Renal chronicity is associated with miR-150, which is negatively regulated by circHLA-C, in patients with lupus nephritis. There is also evidence that circular RNA is involved in acute kidney injury. In these circumstances circular RNA proves to be a novel biomarker and is also used for targeted therapy of kidney disease because its pseudogene can alter DNA composition.
Liver disease
Evidence found that circular RNA plays a role in chronic liver disease and homeostasis
In biology, homeostasis (British English, British also homoeostasis; ) is the state of steady internal physics, physical and chemistry, chemical conditions maintained by organism, living systems. This is the condition of optimal functioning fo ...
regulation leading to liver fibrosis and autoimmune disease by an epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
mechanism.
Cancer
Circular RNA has both positive and negative functions in cancer. For example, ciRS-7 was found to be an oncogene in colorectal cancer tissue that regulates the disease. Overexpression of this ciRS-7 leads to deregulated gene expression leading to malignant phenotypic features. On the other hand, some CircRNAs show positive effects such as circ-ITCH which regulates lung cancer associated with oncogenic sponges miR7 and miR214 and overexpression of circ-ITCH inhibits cell proliferation in lung cancer. From different research studies it has been found that F-circM-9, F-circPR and F-circEA, FcircEA-2 are involved in the development of leukemia and cancer. In osteosarcoma cell circ-0016347 induces tumor and downregulation of caspase-1 target. Another circular RNA hsa_circRNA_002178 leads to breast cancer
Breast cancer is a cancer that develops from breast tissue. Signs of breast cancer may include a Breast lump, lump in the breast, a change in breast shape, dimpling of the skin, Milk-rejection sign, milk rejection, fluid coming from the nipp ...
when it overexpresses and down-regulates COL1A1 protein function. In contrast, silencing of hsa_circRNA_002178 reduced IL-6 and TNF-α production, which inhibited tumor growth and inflammation. Some viruses such as Epstein Barr virus and human papillomavirus
Human papillomavirus infection (HPV infection) is caused by a DNA virus from the ''Papillomaviridae'' family. Many HPV infections cause no symptoms and 90% resolve spontaneously within two years. In some cases, an HPV infection persists and r ...
can encode circular RNAs such as circEBNA_W1_C1 (EBV) and circE7 (HPV) that play a role in oncogenesis in infected individuals. As circRNAs involved in cancer development or regulation process so that it has the potential to use as a biomarker in cancer surveillance and identification process.
Autoimmune disease
Circular RNA has a function in autoimmune disease
An autoimmune disease is a condition that results from an anomalous response of the adaptive immune system, wherein it mistakenly targets and attacks healthy, functioning parts of the body as if they were foreign organisms. It is estimated tha ...
progression acting as a miRNA sponge which regulates DNA methylation, adaptive immune activation, and costimulatory molecule secretion.
Circular RNA in immune response
Circular RNA plays a significant role in immune regulation and induction of T cell responses. circRNA100783 is involved in immunity and senescence of CD8+ T cells. circRNA-003780 and circRNA-010056 also have major roles for macrophage differentiation and polarization.plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
s to express genes of interest.
Circular RNA in Myogenesis regulation
Circular RNA plays an important role in myogenesis
Myogenesis is the formation of skeletal muscle, skeletal muscular tissue, particularly during embryonic development.
Skeletal muscle#Skeletal muscle cells, Muscle fibers generally form through the fusion of precursor cell, precursor myoblasts in ...
mechanisms such as circRBFOX2, circLMO7 acts as a negative regulator and CircSVIL acts as a positive regulator.
Viroids as circular RNAs
Viroid
Viroids are small single-stranded, circular RNAs that are infectious pathogens. Unlike viruses, they have no protein coating. All known viroids are inhabitants of angiosperms (flowering plants), and most cause diseases, whose respective eco ...
s are mostly plant pathogens, which consist of short stretches (a few hundred nucleobases) of highly complementary, circular, single-stranded, and non-coding RNAs without a protein coat. Compared with other infectious plant pathogens, viroids are extremely small in size, ranging from 246 to 467 nucleobases; they thus consist of fewer than 10,000 atoms. In comparison, the genome of the smallest known viruses capable of causing an infection by themselves are around 2,000 nucleobases long.
Circular RNAs in the initial RNA world
It has been proposed that circular RNA genomes which are common in the modern living world may have had their origin at the beginning of life
Life, also known as biota, refers to matter that has biological processes, such as Cell signaling, signaling and self-sustaining processes. It is defined descriptively by the capacity for homeostasis, Structure#Biological, organisation, met ...
.catalytic
Catalysis () is the increase in reaction rate, rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst ...
functions.[ However, self-replication would likely have been favored by a circular structure in which molecular folding was more constrained since circularization would likely interfere with functional folding. Thus the two forms of RNA structure, linear with unconstrained folding allowing the formation of functional domains, and circular with less folding but facilitating smoother replication, may have alternated in early RNA life.][
]
References
{{reflist, 30em
RNA
 In
In  Circular RNAs can be separated into five classes:
Circular RNAs can be separated into five classes: