|
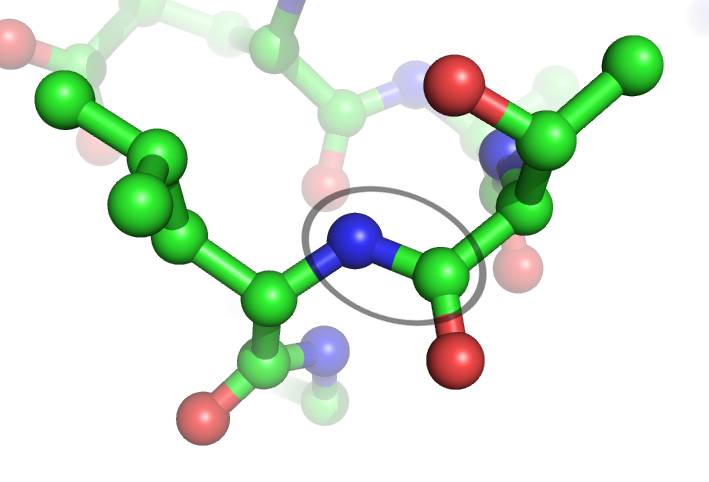
Ubiquitinated
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the 26S proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are essential protein complexes responsible for the degradation of proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are found inside all eukaryotes and archaea, and in some bacteria. In eukaryotes, proteasomes are located both in the nucleus and in the cytoplasm. The proteasomal degradation pathway is essential for many cellular processes, including the cell cycle, the regulation of gene expression, and responses to oxidative stress. The importance of proteolytic degradation inside cells and the role of ubiquitin in proteolytic pathways was acknowledged in the award of the 2004 Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko and Irwin Rose. The core 20S proteasome (blue in the adjacent figure) is a cylindrical, compartmental protein complex of four stacked rings forming a central pore. Each ring is composed of seven individual proteins. The inner two rings a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin C
Polyubiquitin-C is a protein encoded by the ''UBC'' gene in humans. Polyubiquitin-C is one of the sources of ubiquitin, along with UBB, UBA52, and RPS27A. ''UBC'' gene is one of the two stress-regulated polyubiquitin genes (''UBB'' and ''UBC'') in mammals. It plays a key role in maintaining cellular ubiquitin levels under stress conditions. Defects of ''UBC'' gene could lead to mid-gestation embryonic lethality. Structure Gene ''UBC'' gene is located at chromosome 12q24.3, consisting of 2 exons An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence i .... The promoter (genetics), promoter of the ''UBC'' gene contains putative heat shock elements (HSEs), which mediates UBC induction upon stress. ''UBC'' gene differs from ''UBB'' gene in the number of Ub coding units they contain. Nine t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another protein (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin-activating Enzyme
Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which (among other things) can target a protein for degradation via a proteasome. This covalent bond of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms. Many processes such as cell division, immune responses and embryonic development are also regulated by post-translational modification by ubiquitin and ubiquitin-like proteins. Ubiquitination (ubiquitylation) Ubiquitin-activating enzyme (E1) starts the ubiquitination process (Figure 1). The E1 enzyme, along with ATP, binds to the ubiquitin protein. The E1 enzyme then passes the ubiquitin protein to a second protein, called ubiquitin carrier or conjugation protein (E2). The E2 protein complexes with a ubiquitin protein ligase (E3). This ubiquitin protein ligase recognizes which protein needs to be tagged and catalyzes the transfer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Threonine
Threonine (symbol Thr or T) is an amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form when dissolved in water), a carboxyl group (which is in the deprotonated −COO− form when dissolved in water), and a side chain containing a hydroxyl group, making it a polar, uncharged amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Threonine is synthesized from aspartate in bacteria such as ''E. coli''. It is encoded by all the codons starting AC (ACU, ACC, ACA, and ACG). Threonine sidechains are often hydrogen bonded; the most common small motifs formed are based on interactions with serine: ST turns, ST motifs (often at the beginning of alpha helices) and ST staples (usually at the middle of alpha helices). Modifications The threonine residue is susceptible to numerous posttranslational modifications. The hydroxyl side-chain can und ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ester
In chemistry, an ester is a compound derived from an acid (either organic or inorganic) in which the hydrogen atom (H) of at least one acidic hydroxyl group () of that acid is replaced by an organyl group (R). These compounds contain a distinctive functional group. Analogues derived from oxygen replaced by other chalcogens belong to the ester category as well. According to some authors, organyl derivatives of acidic hydrogen of other acids are esters as well (e.g. amides), but not according to the IUPAC. Glycerides are fatty acid esters of glycerol; they are important in biology, being one of the main classes of lipids and comprising the bulk of animal fats and vegetable oils. Lactones are cyclic carboxylic esters; naturally occurring lactones are mainly 5- and 6-membered ring lactones. Lactones contribute to the aroma of fruits, butter, cheese, vegetables like celery and other foods. Esters can be formed from oxoacids (e.g. esters of acetic acid, carbonic acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-terminus
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein. Chemistry Each amino acid has an amine group and a carboxylic group. Amino acids link to one another by peptide bonds which form through a dehydration reaction that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methionine
Methionine (symbol Met or M) () is an essential amino acid in humans. As the precursor of other non-essential amino acids such as cysteine and taurine, versatile compounds such as SAM-e, and the important antioxidant glutathione, methionine plays a critical role in the metabolism and health of many species, including humans. Methionine is also involved in angiogenesis and various processes related to DNA transcription, epigenetic expression, and gene regulation. Methionine was first isolated in 1921 by John Howard Mueller. It is Genetic code, encoded by the codon AUG. It was named by Satoru Odake in 1925, as an abbreviation of its structural description 2-amino-4-(methylthio)butanoic acid. Biochemical details Methionine (abbreviated as Met or M; encoded by the codon AUG) is an α-amino acid that is used in the biosynthesis of proteins. It contains a carboxyl group (which is in the deprotonated −COO− form under biological pH conditions), an amino group (which is in the proton ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a '' dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
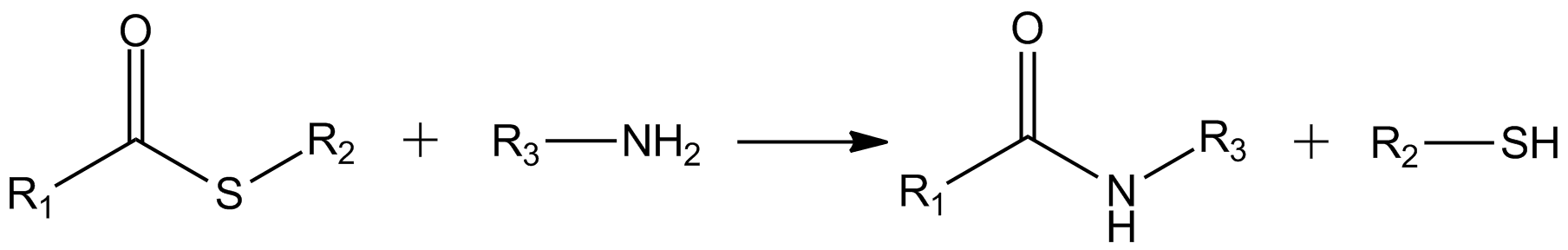
Thioester
In organic chemistry, thioesters are organosulfur compounds with the molecular structure . They are analogous to carboxylate esters () with the sulfur in the thioester replacing oxygen in the carboxylate ester, as implied by the thio- prefix. They are the product of esterification of a carboxylic acid () with a thiol (). In biochemistry, the best-known thioesters are derivatives of coenzyme A, e.g., acetyl-CoA.Matthys J. Janssen "Carboxylic Acids and Esters" in PATAI's Chemistry of Functional Groups: Carboxylic Acids and Esters, Saul Patai, Ed. John Wiley, 1969, New York: pp. 705–764. The R and R' represent organyl groups, or H in the case of R. Synthesis One route to thioesters involves the reaction of an acid chloride with an alkali metal salt of a thiol: : Another common route entails the displacement of halides by the alkali metal salt of a thiocarboxylic acid. For example, thioacetate esters are commonly prepared by alkylation of potassium thioacetate: : Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
One-letter Amino Acid Notation
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups ( alpha- , beta- , gamma- amino acids, etc.); other categories relate to polarity, ionization, and side-chain group type (aliphatic, acyclic, aromatic, polar, etc.). In the form of proteins, amino-acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life on Earth and its emergence. Amino acids are formally named by the IUPAC- IUBMB Joint Commissio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |