|
Torsion Angle
In stereochemistry, a torsion angle is defined as a particular example of a dihedral angle, describing the geometric relation of two parts of a molecule joined by a chemical bond. Every set of three non-colinear atoms of a molecule defines a half-plane. As explained above, when two such half-planes intersect (i.e., a set of four consecutively-bonded atoms), the angle between them is a dihedral angle. Dihedral angles are used to specify the Conformational isomerism, molecular conformation. Stereochemical arrangements corresponding to angles between 0° and ±90° are called ''syn'' (s), those corresponding to angles between ±90° and 180° ''anti'' (a). Similarly, arrangements corresponding to angles between 30° and 150° or between −30° and −150° are called ''clinal'' (c) and those between 0° and ±30° or ±150° and 180° are called ''periplanar'' (p). The two types of terms can be combined so as to define four ranges of angle; 0° to ±30° synperiplanar (sp); 30° to ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
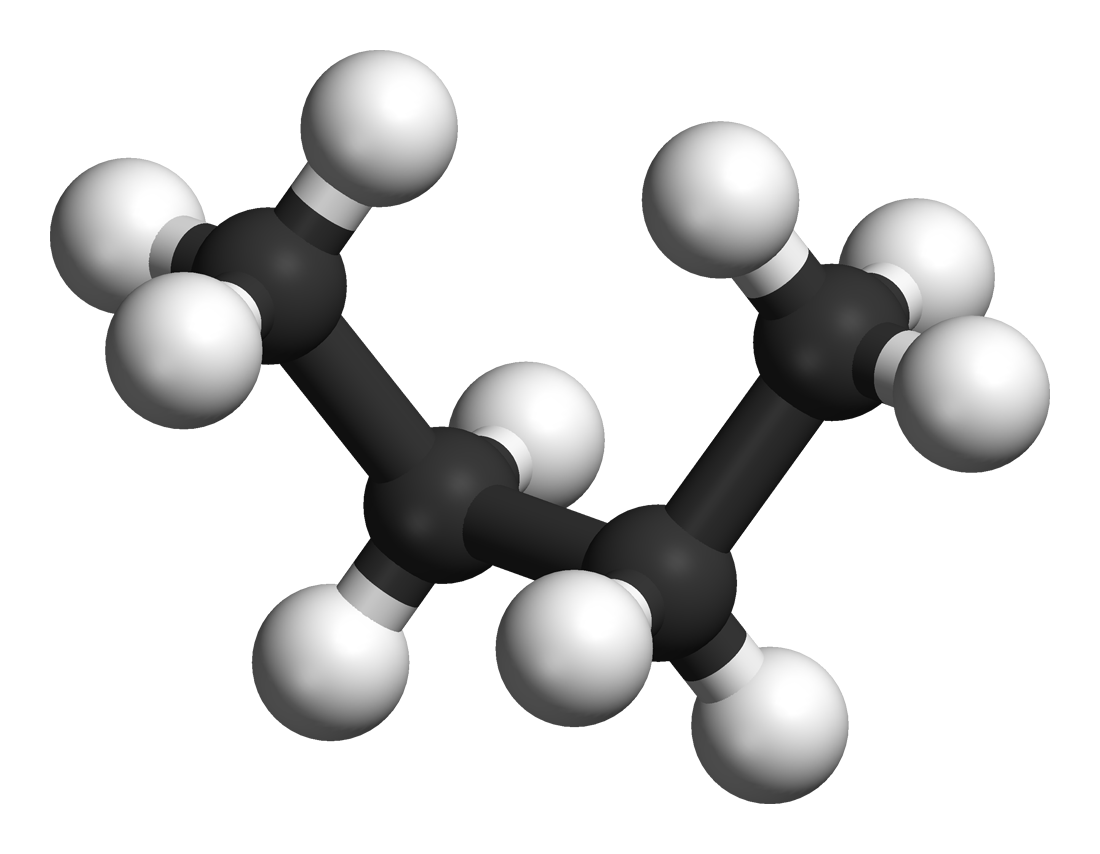
Dihedral Angles Of Butane , a geometric shape
{{ ...
Dihedral or polyhedral may refer to: * Dihedral angle, the angle between two mathematical planes * Dihedral (aeronautics), the upward angle of a fixed-wing aircraft's wings where they meet at the fuselage, dihedral ''effect'' of an aircraft, longitudinal dihedral angle of a fixed-wing aircraft * Dihedral group, the group of symmetries of the ''n''-sided polygon in abstract algebra ** Also Dihedral symmetry in three dimensions * Dihedral kite, also known as a bowed kite * Dihedral doors, also known as butterfly doors * Dihedral prime, also known as a dihedral calculator prime * In rock climbing, an inside corner of rock See also * Anhedral (other) * Euhedral, a crystal structure * Polyhedron In geometry, a polyhedron (: polyhedra or polyhedrons; ) is a three-dimensional figure with flat polygonal Face (geometry), faces, straight Edge (geometry), edges and sharp corners or Vertex (geometry), vertices. The term "polyhedron" may refer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Backbone PhiPsiOmega Drawing
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Backbone-dependent Rotamer Library
In biochemistry, a backbone-dependent rotamer library provides the frequencies, mean dihedral angles, and standard deviations of the discrete conformations (known as rotamers) of the amino acid Side_chain#Biochemistry, side chains in proteins as a function of the protein backbone, backbone dihedral angles φ and ψ of the Ramachandran plot, Ramachandran map. By contrast, backbone-independent rotamer libraries express the frequencies and mean dihedral angles for all side chains in proteins, regardless of the backbone conformation of each residue type. Backbone-dependent rotamer libraries have been shown to have significant advantages over backbone-independent rotamer libraries, principally when used as an energy term, by speeding up search times of side-chain packing algorithms used in protein structure prediction and protein design. History The first backbone-dependent rotamer library was developed in 1993 by Roland Dunbrack and Martin Karplus to assist the prediction of the Carte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the protonated form (NH2+) under biological conditions, while the carboxyl group is in the deprotonated −COO− form. The "side chain" from the α carbon connects to the nitrogen forming a pyrrolidine loop, classifying it as a aliphatic amino acid. It is non-essential in humans, meaning the body can synthesize it from the non-essential amino acid L-glutamate. It is encoded by all the codons starting with CC (CCU, CCC, CCA, and CCG). Proline is the only proteinogenic amino acid which is a secondary amine, as the nitrogen atom is attached both to the α-carbon and to a chain of three carbons that together form a five-membered ring. History and etymology Proline was first isolated in 1900 by Richard Willstätter who obtained the amino a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Geometric Isomerism
Geometry (; ) is a branch of mathematics concerned with properties of space such as the distance, shape, size, and relative position of figures. Geometry is, along with arithmetic, one of the oldest branches of mathematics. A mathematician who works in the field of geometry is called a '' geometer''. Until the 19th century, geometry was almost exclusively devoted to Euclidean geometry, which includes the notions of point, line, plane, distance, angle, surface, and curve, as fundamental concepts. Originally developed to model the physical world, geometry has applications in almost all sciences, and also in art, architecture, and other activities that are related to graphics. Geometry also has applications in areas of mathematics that are apparently unrelated. For example, methods of algebraic geometry are fundamental in Wiles's proof of Fermat's Last Theorem, a problem that was stated in terms of elementary arithmetic, and remained unsolved for several centuries. During t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
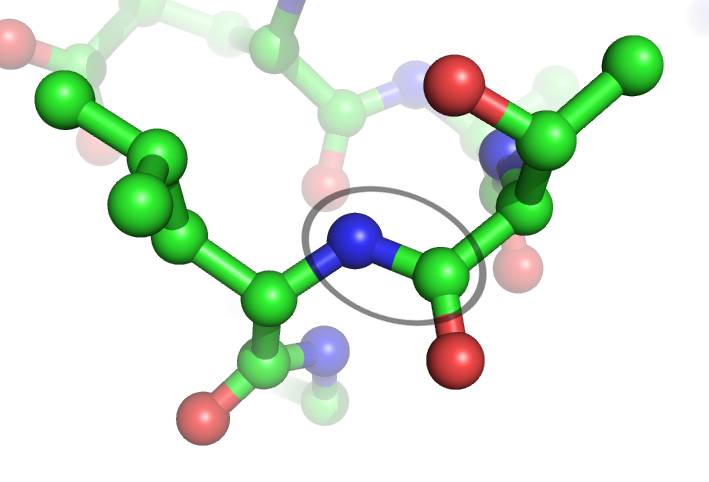
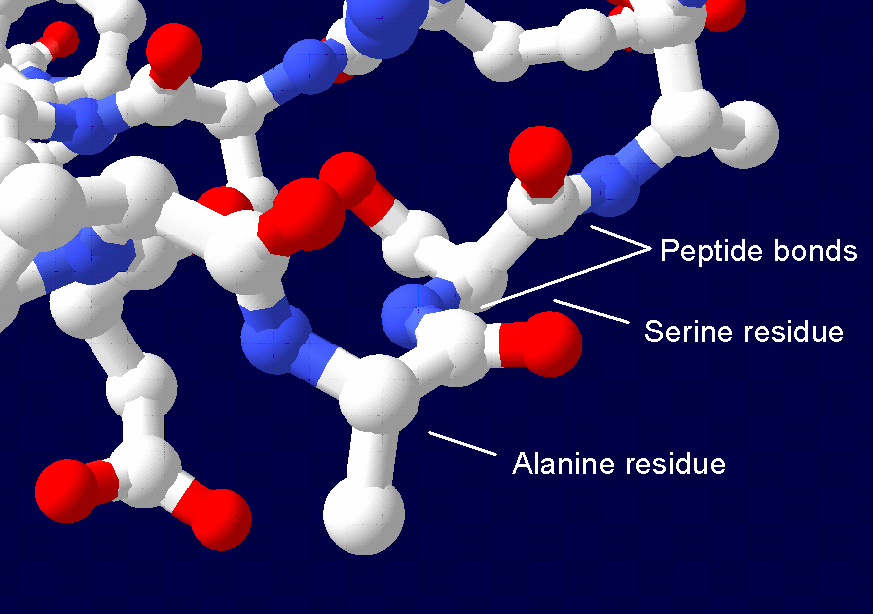
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a '' dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid monomer may also be called a ''residue'', which indicates a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions, such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensiona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 appear in the genetic code of life. Amino acids can be classified according to the locations of the core structural functional groups ( alpha- , beta- , gamma- amino acids, etc.); other categories relate to polarity, ionization, and side-chain group type ( aliphatic, acyclic, aromatic, polar, etc.). In the form of proteins, amino-acid '' residues'' form the second-largest component (water being the largest) of human muscles and other tissues. Beyond their role as residues in proteins, amino acids participate in a number of processes such as neurotransmitter transport and biosynthesis. It is thought that they played a key role in enabling life on Earth and its emergence. Amino acids are formally named by the IUPAC- IUBMB Joint Commi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ramachandran Plot
In biochemistry, a Ramachandran plot (also known as a Rama plot, a Ramachandran diagram or a �,ψplot), originally developed in 1963 by G. N. Ramachandran, C. Ramakrishnan, and V. Sasisekharan, is a way to visualize energetically allowed regions for backbone dihedral angles (also called as torsional angles, phi and psi angles) ψ against φ of amino acid residues in protein structure. The figure on the left illustrates the definition of the φ and ψ backbone dihedral angles (called φ and φ' by Ramachandran). The ω angle at the peptide bond is normally 180°, since the partial-double-bond character keeps the peptide bond planar. The figure in the top right shows the allowed φ,ψ backbone conformational regions from the Ramachandran et al. 1963 and 1968 hard-sphere calculations: full radius in solid outline, reduced radius in dashed, and relaxed tau (N-Cα-C) angle in dotted lines. Because dihedral angle values are circular and 0° is the same as 360°, the edges of the R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Newman Projection
A Newman projection is a drawing that helps visualize the 3-dimensional structure of a molecule. This projection most commonly sights down a carbon-carbon bond, making it a very useful way to visualize the stereochemistry of alkanes. A Newman projection visualizes the Conformational isomerism, conformation of a chemical bond from front to back, with the front atom represented by the intersection of three lines (a dot) and the back atom as a circle. The front atom is called ''proximal'', while the back atom is called ''distal''. This type of representation clearly illustrates the specific dihedral angle between the proximal and distal atoms. This projection is named after American chemist Melvin Spencer Newman, who introduced it in 1952 as a partial replacement for Fischer projections, which are unable to represent conformations and thus conformers properly.Newman, MS. ''Record. Chem. Progr. (Kresge-Hooker Sci. Lib.) 1952,'' 13'', 111'' This diagram style is an alternative to a sa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |