|
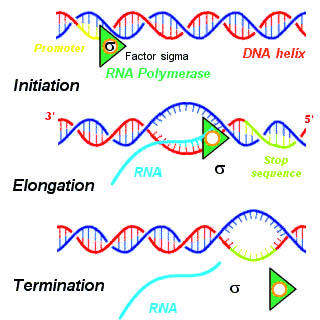
Termination Signal
In molecular biology, a termination signal is a sequence that signals the end of transcription or translation. Termination signals are found at the end of the part of the chromosome being transcribed during transcription of mRNA. Termination signals bring a stop to transcription, ensuring that only gene-encoding parts of the chromosome are transcribed. Transcription begins at the promoter when RNA polymerase, an enzyme that facilitates transcription of DNA into mRNA, binds to a promoter, unwinds the helical structure of the DNA, and uses the single-stranded DNA as a template to synthesize RNA. Once RNA polymerase reaches the termination signal, transcription is terminated. In bacteria, there are two main types of termination signals: intrinsic and factor-dependent terminators. In the context of translation, a termination signal is the stop codon on the mRNA that elicits the release of the growing peptide from the ribosome. Termination signals play an important role in regulati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription
Transcription refers to the process of converting sounds (voice, music etc.) into letters or musical notes, or producing a copy of something in another medium, including: Genetics * Transcription (biology), the copying of DNA into RNA, often the first step in gene expression ** Abortive transcription, the generation of very short RNA transcripts which are not used and rapidly degraded ** Bacterial transcription, the generation of RNA transcripts of the genetic material in bacteria ** Eukaryotic transcription, the process of copying the genetic information stored in DNA into RNA in eukaryotes ** Reverse transcription, the process of copying the genetic information stored in RNA into DNA in viruses ** ''Transcription'' (journal), an academic journal about genetics ** Transcription factor, a protein that controls the rate of transcription of genetic information from DNA to messenger RNA Music * Transcription (music), notating, converting musical sound into visual musical notes (fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsic Termination
Intrinsic, or rho-independent termination, is a process to signal the end of transcription and release the newly constructed RNA molecule. In bacteria such as ''E. coli'', transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil (rU) and adenine (dA) are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is complementary and pairs to either thymine in DNA or uracil in RNA. In cells adenine, as an independent molecule, is rare. It is almost always covalent bond, covalently bound to become a part of a larger biomolecule. Adenine has a central role in cellular respiration. It is part of adenosine triphosphate which provides the energy that drives and supports most activities in living cell (biology), cells, such as Protein biosynthesis, protein synthesis, chemical synthesis, muscle contraction, and nerve impulse propagation. In respiration it also participates as part of the cofactor (biochemistry), cofactors nicotinamide adenine dinucleotide, flavin adenine dinucleotide, and Coenzyme A. It is also part of adenosine, adenosine monophosphate, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Purine
Purine is a heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature. Dietary sources Purines are found in high concentration in meat and meat products, especially internal organs, such as liver and kidney, and in various seafoods, high-fructose beverages, alcohol, and yeast products. Examples of high-purine food sources include anchovies, sardines, liver, beef, kidneys, brains, monkfish, dried mackerel, and shrimp. Foods particularly rich in hypoxanthine, adenine, and guanine lead to higher blood levels of uric acid. Foods having more than 200 mg of hypoxanthine per 100 g, particularly animal and fish meats containing hypoxanthine as more than 50% of total purines, are more likely to increase uri ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T7 Promoter
T7 RNA Polymerase is an RNA polymerase from the T7 bacteriophage that catalyzes the formation of RNA from DNA in the 5'→ 3' direction. Activity T7 polymerase is extremely promoter-specific and transcribes only DNA downstream of a T7 promoter. The T7 polymerase also requires a double stranded DNA template and Mg2+ ion as cofactor for the synthesis of RNA. It has a very low error rate. T7 polymerase has a molecular weight of 99 kDa. Promoter The promoter is recognized for binding and initiation of the transcription. The consensus in T7 and related phages is: 5' * 3' T7 TAATACGACTCACTATAGGGAGA T3 AATTAACCCTCACTAAAGGGAGA K11 AATTAGGGCACACTATAGGGAGA SP6 ATTTACGACACACTATAGAAGAA bind------------ -----------init Transcription begins at the asterisk-marked guanine. Structure T7 polymerase has been crystallised in several forms and the structures placed in the PDB. These explain how T7 polymerase binds to DNA and trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and archaea; however plasmids are sometimes present in and eukaryotic organisms as well. Plasmids often carry useful genes, such as those involved in antibiotic resistance, virulence, secondary metabolism and bioremediation. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain additional genes for special circumstances. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet by various vendors ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Recombinant Proteins
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome. Recombinant DNA is the general name for a piece of DNA that has been created by combining two or more fragments from different sources. Recombinant DNA is possible because DNA molecules from all organisms share the same chemical structure, differing only in the nucleotide sequence. Recombinant DNA molecules are sometimes called chimeric DNA because they can be made of material from two different species like the mythical chimera. rDNA technology uses palindromic sequences and leads to the production of sticky and blunt ends. The DNA sequences used in the construction of recombinant DNA molecules can originate from any species. For example, plant DNA can be joined to bacterial DNA, or human DNA can be joined with fungal D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
T7 RNA Polymerase
T7 RNA Polymerase is an RNA polymerase from the T7 bacteriophage that catalyzes the formation of RNA from DNA in the 5'→ 3' direction. Activity T7 polymerase is extremely promoter-specific and transcribes only DNA downstream of a T7 promoter. The T7 polymerase also requires a double stranded DNA template and Mg2+ ion as cofactor for the synthesis of RNA. It has a very low error rate. T7 polymerase has a molecular weight of 99 kDa. Promoter The promoter is recognized for binding and initiation of the transcription. The consensus in T7 and related phages is: 5' * 3' T7 TAATACGACTCACTATAGGGAGA T3 AATTAACCCTCACTAAAGGGAGA K11 AATTAGGGCACACTATAGGGAGA SP6 ATTTACGACACACTATAGAAGAA bind------------ -----------init Transcription begins at the asterisk-marked guanine. Structure T7 polymerase has been crystallised in several forms and the structures placed in the PDB. These explain how T7 polymerase binds to DNA and trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antitermination
{{short description, Genetic transcription mechanism in prokaryotes In molecular biology, antitermination is the prokaryotic cell's aid to fix premature termination during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal. Antitermination is used by some phages to regulate progression from one stage of gene expression to the next. The lambda gene N, codes for an antitermination protein (pN) that is necessary to allow RNA polymerase to read through the terminators located at the ends of the immediate early genes. Another antitermination protein, pQ, is required later in phage infection. pN and pQ act on RNA polymerase as it passes specific sites. These sites are l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rho Family Of GTPases
The Rho family of GTPases is a family of small (~21 kDa) signaling G proteins, and is a subfamily of the Ras superfamily. The members of the Rho GTPase family have been shown to regulate many aspects of intracellular actin dynamics, and are found in all eukaryotic kingdoms, including yeasts and some plants. Three members of the family have been studied in detail: Cdc42, Rac1, and RhoA. All G proteins are "molecular switches", and Rho proteins play a role in organelle development, cytoskeletal dynamics, cell movement, and other common cellular functions. History Identification of the Rho family of GTPases began in the mid-1980s. The first identified Rho member was RhoA, isolated serendipitously in 1985 from a low stringency cDNA screening. Rac1 and Rac2 were identified next, in 1989 followed by Cdc42 in 1990. Eight additional mammalian Rho members were identified from biological screenings until the late 1990s, a turning point in biology where availability of complete genome sequenc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleobase
Nucleotide bases (also nucleobases, nitrogenous bases) are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA). Five nucleobases— adenine (A), cytosine (C), guanine (G), thymine (T), and uracil (U)—are called ''primary'' or ''canonical''. They function as the fundamental units of the genetic code, with the bases A, G, C, and T being found in DNA while A, G, C, and U are found in RNA. Thymine and uracil are distinguished by merely the presence or absence of a methyl group on the fifth carbon (C5) of these heterocyclic six-membered rings. In addition, some viruses have aminoadenine (Z) instead of adenine. It differs in having an extra amine group, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine (T). Uracil is a demethylated form of thymine. Uracil is a common and naturally occurring pyrimidine derivative. The name "uracil" was coined in 1885 by the German chemist Robert Behrend, who was attempting to synthesize derivatives of uric acid. Originally discovered in 1900 by Alberto Ascoli, it was isolated by hydrolysis of yeast nuclein; it was also found in bovine thymus and spleen, herring sperm, and wheat Cereal germ, germ. It is a planar, unsaturated compound that has the ability to absorb light. Uracil that was formed extraterrestrially has been detected in the Murchison meteorite, in near-Earth asteroid 162173 Ryugu, Ryugu, and possibly on the surface of th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |