|
Tandem Mass Tag
In analytical chemistry, a tandem mass tag (TMT) is a chemical label that facilitates sample multiplexing in mass spectrometry (MS)-based quantification and identification of biological macromolecules such as proteins, peptides and nucleic acids. TMT belongs to a family of reagents referred to as isobaric mass tags which are a set of molecules with the same mass, but yield reporter ions of differing mass after fragmentation. The relative ratio of the measured reporter ions represents the relative abundance of the tagged molecule, although ion suppression has a detrimental effect on accuracy. Despite these complications, TMT-based proteomics has been shown to afford higher precision than label-free quantification. In addition to aiding in protein quantification, TMT tags can also increase the detection sensitivity of certain highly hydrophilic analytes, such as phosphopeptides, in RPLC-MS analyses. Versions There are currently six varieties of TMT available: TMTzero, a non-isot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Analytical Chemistry
Analytical skill, Analytical chemistry studies and uses instruments and methods to Separation process, separate, identify, and Quantification (science), quantify matter. In practice, separation, identification or quantification may constitute the entire analysis or be combined with another method. Separation isolates analytes. Qualitative inorganic analysis, Qualitative analysis identifies analytes, while Quantitative analysis (chemistry), quantitative analysis determines the numerical amount or concentration. Analytical chemistry consists of classical, wet chemistry, wet chemical methods and modern analytical techniques. Classical qualitative methods use separations such as Precipitation (chemistry), precipitation, Extraction (chemistry), extraction, and distillation. Identification may be based on differences in color, odor, melting point, boiling point, solubility, radioactivity or reactivity. Classical quantitative analysis uses mass or volume changes to quantify amount. Ins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Mass
The molecular mass () is the mass of a given molecule, often expressed in units of daltons (Da). Different molecules of the same compound may have different molecular masses because they contain different isotopes of an element. The derived quantity relative molecular mass is the unitless ratio of the mass of a molecule to the atomic mass constant (which is equal to one dalton). The molecular mass and relative molecular mass are distinct from but related to the ''molar mass''. The molar mass is defined as the mass of a given substance divided by the amount of the substance, and is expressed in grams per mole (g/mol). That makes the molar mass an ''average'' of many particles or molecules (weighted by abundance of the isotopes), and the molecular mass the mass of one specific particle or molecule. The molar mass is usually the more appropriate quantity when dealing with macroscopic (weigh-able) quantities of a substance. The definition of molecular weight is most authoritat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemistry Detection Methods
Biochemistry, or biological chemistry, is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology, and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all areas of the life sciences are being uncovered and developed through biochemical methodology and research. Voet (2005), p. 3. Biochemistry focuses on understanding the chemical basis that allows biological molecules to give rise to the processes that occur within living cells and between cells, Karp (2009), p. 2. in turn relating greatly to the understanding of tissues and organs as well as organism structure and function. Miller (2012). p. 62. Biochemistry is closely related to molecular biology, the study of the molecular mechanisms of biological phenomena.Astbury ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mascot (software)
Mascot is a software search engine that uses mass spectrometry data to identify proteins from peptide sequence databases. Mascot is widely used by research facilities around the world. Mascot uses a probabilistic scoring algorithm for protein identification that was adapted from the MOWSE algorithm. Mascot is freely available to use on the website of Matrix Science. A license is required for in-house use where more features can be incorporated. History means MOWSE was one of the first algorithms developed for protein identification using peptide mass fingerprinting. It was originally developed in 1993 as a collaboration between Darryl Pappin of the Imperial Cancer Research Fund (ICRF) and Alan Bleasby of the Science and Engineering Research Council (SERC). MOWSE stood apart from other protein identification algorithms in that it produced a probability-based score for identification. It was also the first to take into account the non-uniform distribution of peptide sizes, caused ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mass Spectrometry Software
Mass spectrometry software is used for data acquisition, analysis, or representation in mass spectrometry. Proteomics software In protein mass spectrometry, tandem mass spectrometry (also known as MS/MS or MS2) experiments are used for protein/peptide identification. Peptide identification algorithms fall into two broad classes: database search and ''de novo'' search. The former search takes place against a database containing all amino acid sequences assumed to be present in the analyzed sample. In contrast, the latter infers Protein primary structure, peptide sequences without knowledge of genomic data. Database search algorithms De novo sequencing algorithms De novo peptide sequencing algorithms are, in general, based on the approach proposed in Bartels ''et al.'' (1990). Homology searching algorithms MS/MS peptide quantification Other software See also * Mass spectrometry data format: for a list of mass spectrometry data viewers and format converters. * List of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Structure
A chemical structure of a molecule is a spatial arrangement of its atoms and their chemical bonds. Its determination includes a chemist's specifying the molecular geometry and, when feasible and necessary, the electronic structure of the target molecule or other solid. Molecular geometry refers to the spatial arrangement of atoms in a molecule and the chemical bonds that hold the atoms together and can be represented using structural formulae and by molecular models; complete electronic structure descriptions include specifying the occupation of a molecule's molecular orbitals. Structure determination can be applied to a range of targets from very simple molecules (e.g., diatomic oxygen or nitrogen) to very complex ones (e.g., such as protein or DNA). Background Theories of chemical structure were first developed by August Kekulé, Archibald Scott Couper, and Aleksandr Butlerov, among others, from about 1858. These theories were first to state that chemical compounds are not a ran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
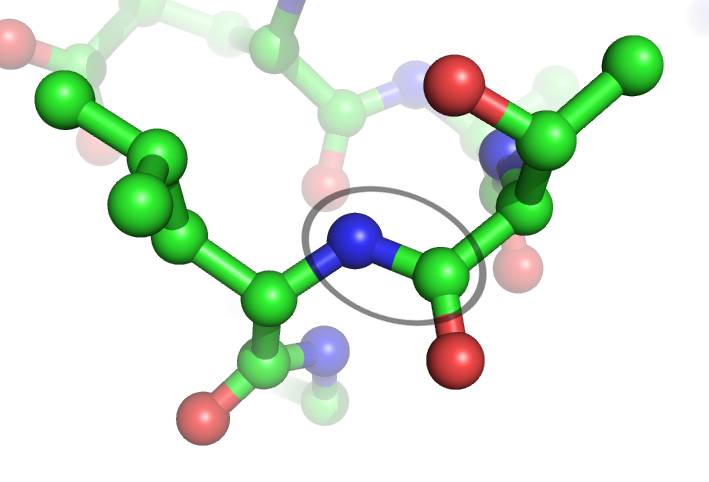
Peptide Bond
In organic chemistry, a peptide bond is an amide type of covalent chemical bond linking two consecutive alpha-amino acids from C1 (carbon number one) of one alpha-amino acid and N2 (nitrogen number two) of another, along a peptide or protein chain. It can also be called a eupeptide bond to distinguish it from an isopeptide bond, which is another type of amide bond between two amino acids. Synthesis When two amino acids form a '' dipeptide'' through a ''peptide bond'', it is a type of condensation reaction. In this kind of condensation, two amino acids approach each other, with the non-side chain (C1) carboxylic acid moiety of one coming near the non-side chain (N2) amino moiety of the other. One loses a hydrogen and oxygen from its carboxyl group (COOH) and the other loses a hydrogen from its amino group (NH2). This reaction produces a molecule of water (H2O) and two amino acids joined by a peptide bond (−CO−NH−). The two joined amino acids are called a dipeptide. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandem Mass Spectrometry
Tandem mass spectrometry, also known as MS/MS or MS2, is a technique in instrumental analysis where two or more stages of analysis using one or more mass analyzer are performed with an additional reaction step in between these analyses to increase their abilities to analyse chemical samples. A common use of tandem MS is the analysis of biomolecules, such as proteins and peptides. The molecules of a given sample are ionized and the first spectrometer (designated MS1) separates these ions by their mass-to-charge ratio (often given as m/z or m/Q). Ions of a particular m/z-ratio coming from MS1 are selected and then made to split into smaller fragment ions, e.g. by collision-induced dissociation, ion-molecule reaction, or photodissociation. These fragments are then introduced into the second mass spectrometer (MS2), which in turn separates the fragments by their m/z-ratio and detects them. The fragmentation step makes it possible to identify and separate ions that have very simil ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
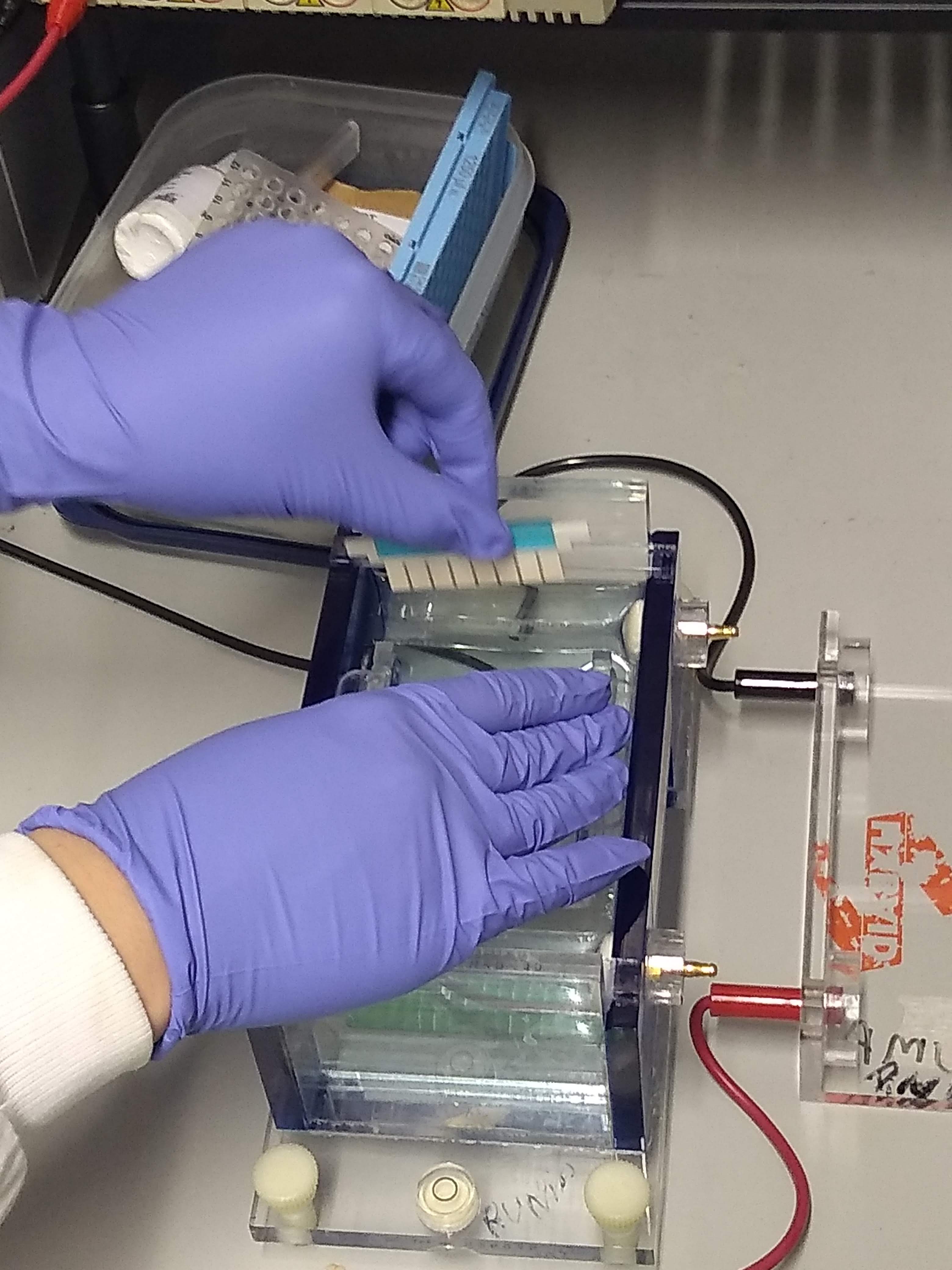
Gel Electrophoresis
Gel electrophoresis is an electrophoresis method for separation and analysis of biomacromolecules (DNA, RNA, proteins, etc.) and their fragments, based on their size and charge through a gel. It is used in clinical chemistry to separate proteins by charge or size (IEF agarose, essentially size independent) and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments, or to separate proteins by charge. Nucleic acid molecules are separated by applying an electric field to move the negatively charged molecules through a gel matrix of agarose, polyacrylamide, or other substances. Shorter molecules move faster and migrate farther than longer ones because shorter molecules migrate more easily through the pores of the gel. This phenomenon is called sieving. Proteins are separated by the charge in agarose because the pores of the gel are too large to sieve proteins. Gel electrophoresi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatography
In chemical analysis, chromatography is a laboratory technique for the Separation process, separation of a mixture into its components. The mixture is dissolved in a fluid solvent (gas or liquid) called the ''mobile phase'', which carries it through a system (a column, a capillary tube, a plate, or a sheet) on which a material called the ''stationary phase'' is fixed. Because the different constituents of the mixture tend to have different affinities for the stationary phase and are retained for different lengths of time depending on their interactions with its surface sites, the constituents travel at different apparent velocities in the mobile fluid, causing them to separate. The separation is based on the differential partitioning between the mobile and the stationary phases. Subtle differences in a compound's partition coefficient result in differential retention on the stationary phase and thus affect the separation. Chromatography may be ''preparative'' or ''analytical' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isotopes
Isotopes are distinct nuclear species (or ''nuclides'') of the same chemical element. They have the same atomic number (number of protons in their nuclei) and position in the periodic table (and hence belong to the same chemical element), but different nucleon numbers (mass numbers) due to different numbers of neutrons in their nuclei. While all isotopes of a given element have similar chemical properties, they have different atomic masses and physical properties. The term isotope is derived from the Greek roots isos ( ἴσος "equal") and topos ( τόπος "place"), meaning "the same place"; thus, the meaning behind the name is that different isotopes of a single element occupy the same position on the periodic table. It was coined by Scottish doctor and writer Margaret Todd in a 1913 suggestion to the British chemist Frederick Soddy, who popularized the term. The number of protons within the atom's nucleus is called its atomic number and is equal to the number of el ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiplexing
In telecommunications and computer networking, multiplexing (sometimes contracted to muxing) is a method by which multiple analog or digital signals are combined into one signal over a shared medium. The aim is to share a scarce resource—a physical transmission medium. For example, in telecommunications, several telephone calls may be carried using one wire. Multiplexing originated in telegraphy in the 1870s, and is now widely applied in communications. In telephony, George Owen Squier is credited with the development of telephone carrier multiplexing in 1910. The multiplexed signal is transmitted over a communication channel such as a cable. The multiplexing divides the capacity of the communication channel into several logical channels, one for each message signal or data stream to be transferred. A reverse process, known as demultiplexing, extracts the original channels on the receiver end. A device that performs the multiplexing is called a multiplexer (MUX), and a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |