|
Hypodysfibrinogenemia
Hypodysfibrinogenemia, also termed congenital hypodysfibrinogenemia, is a rare hereditary fibrinogen disorder cause by mutations in one or more of the genes that encode a factor critical for blood clotting, fibrinogen. These mutations result in the production and circulation at reduced levels of fibrinogen at least some of which is dysfunctional. Hypodysfibrinogenemia exhibits reduced penetrance, i.e. only some family members with the mutated gene develop symptoms. The disorder is similar to a form of dysfibrinogenemia termed congenital dysfibrinogenemia. However, congenital dysfibrinogenemia differs form hypodysfibrinogenemia in four ways. Congenital dysfibrinogenemia involves: the circulation at normal levels of fibrinogen at least some of which is dysfunctional; a different set of causative gene mutations; a somewhat different mix of clinical symptoms; and a much lower rate of penetrance. Hypodysfibrinogenemia causes episodes of pathological bleeding and thrombosis due not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fibrinogen
Fibrinogen (factor I) is a glycoprotein complex, produced in the liver, that circulates in the blood of all vertebrates. During tissue and vascular injury, it is converted enzymatically by thrombin to fibrin and then to a fibrin-based blood clot. Fibrin clots function primarily to occlude blood vessels to stop bleeding. Fibrin also binds and reduces the activity of thrombin. This activity, sometimes referred to as antithrombin I, limits clotting. Fibrin also mediates blood platelet and endothelial cell spreading, tissue fibroblast proliferation, capillary tube formation, and angiogenesis and thereby promotes revascularization and wound healing. Reduced and/or dysfunctional fibrinogens occur in various congenital and acquired human fibrinogen-related disorders. These disorders represent a group of rare conditions in which individuals may present with severe episodes of pathological bleeding and thrombosis; these conditions are treated by supplementing blood fibrinoge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Fibrinogen Disorders
Fibrinogen disorders are a set of hereditary or acquired abnormalities in the quantity and/or quality of circulating fibrinogens. The disorders may lead to pathological bleeding and/or blood clotting or the deposition of fibrinogen in the liver, kidneys, or other organs and tissues. These disorders include: * Congenital afibrinogenemia, an inherited blood disorder in which blood does not clot normally due to the lack of fibrinogen; the disorder causes abnormal bleeding and thrombosis. * Congenital hypofibrinogenemia, an inherited disorder in which blood may not clot normally due to reduced levels of fibrinogen; the disorder may cause abnormal bleeding and thrombosis. * Fibrinogen storage disease, a form of congenital hypofibrinogenemia in which specific hereditary mutations in fibrinogen cause it to accumulate in, and damage, liver cells. The disorder may lead to abnormal bleeding and thrombosis but also to cirrhosis. * Congenital dysfibrinogenemia, an inherited disorder in which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mutations
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics (phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutation is the ultimate source of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deep Vein Thrombosis
Deep vein thrombosis (DVT) is a type of venous thrombosis involving the formation of a blood clot in a deep vein, most commonly in the legs or pelvis. A minority of DVTs occur in the arms. Symptoms can include pain, swelling, redness, and enlarged veins in the affected area, but some DVTs have no symptoms. The most common life-threatening concern with DVT is the potential for a clot to embolize (detach from the veins), travel as an embolus through the right side of the heart, and become lodged in a pulmonary artery that supplies blood to the lungs. This is called a pulmonary embolism (PE). DVT and PE comprise the cardiovascular disease of venous thromboembolism (VTE). About two-thirds of VTE manifests as DVT only, with one-third manifesting as PE with or without DVT. The most frequent long-term DVT complication is post-thrombotic syndrome, which can cause pain, swelling, a sensation of heaviness, itching, and in severe cases, ulcers. Recurrent VTE occurs in about 30% ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Compound Heterozygosity
In medical genetics, compound heterozygosity is the condition of having two or more heterogeneous recessive alleles at a particular locus that can cause genetic disease in a heterozygous state; that is, an organism is a compound heterozygote when it has two recessive alleles for the same gene, but with those two alleles being different from each other (for example, both alleles might be mutated but at different locations). Compound heterozygosity reflects the diversity of the mutation base for many autosomal recessive genetic disorders; mutations in most disease-causing genes have arisen many times. This means that many cases of disease arise in individuals who have two unrelated alleles, who technically are heterozygotes, but both the alleles are defective. These disorders are often best known in some classic form, such as the homozygous recessive case of a particular mutation that is widespread in some population. In its compound heterozygous forms, the disease may have lower pe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homozygous
Zygosity (the noun, zygote, is from the Greek "yoked," from "yoke") () is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism. Most eukaryotes have two matching sets of chromosomes; that is, they are diploid. Diploid organisms have the same loci on each of their two sets of homologous chromosomes except that the sequences at these loci may differ between the two chromosomes in a matching pair and that a few chromosomes may be mismatched as part of a chromosomal sex-determination system. If both alleles of a diploid organism are the same, the organism is homozygous at that locus. If they are different, the organism is heterozygous at that locus. If one allele is missing, it is hemizygous, and, if both alleles are missing, it is nullizygous. The DNA sequence of a gene often varies from one individual to another. These gene variants are called alleles. While some ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
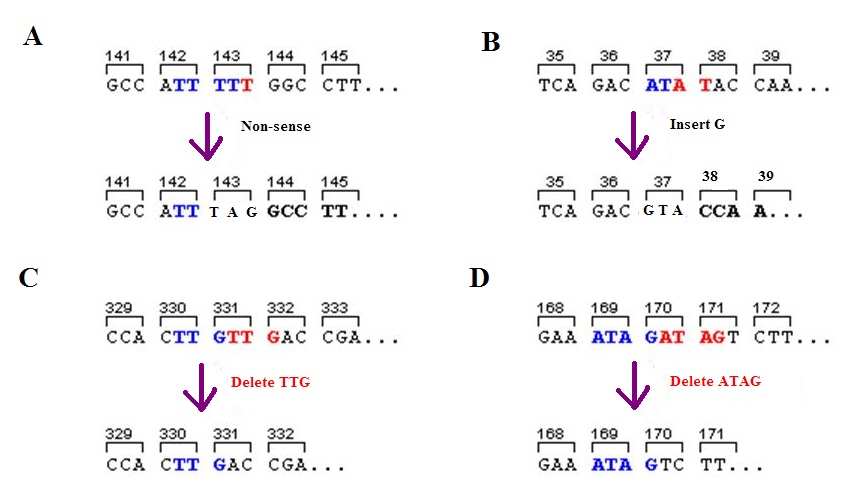
Frameshift Mutation
A frameshift mutation (also called a framing error or a reading frame shift) is a genetic mutation caused by indels (gene insertion, insertions or genetic deletion, deletions) of a number of nucleotides in a DNA sequence that is not divisible by three. Due to the triplet nature of gene expression by codons, the insertion or deletion can change the reading frame (the grouping of the codons), resulting in a completely different Translation (genetics), translation from the original. The earlier in the sequence the deletion or insertion occurs, the more altered the protein. A frameshift mutation is not the same as a single-nucleotide polymorphism in which a nucleotide is replaced, rather than inserted or deleted. A frameshift mutation will in general cause the reading of the codons after the mutation to code for different amino acids. The frameshift mutation will also alter the first stop codon ("UAA", "UGA" or "UAG") encountered in the sequence. The polypeptide being created could be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nonsense Mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a ''nonsense codon'' in the transcribed mRNA, and in leading to a truncated, incomplete, and usually nonfunctional protein product. The functional effect of a nonsense mutation depends on the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations. Missense mutations differ from nonsense mutations since they are point mutations that exhibit a single nucleotide change to cause substitution of a different amino acid. A nonsense mutation also differs from a nonstop mutation, which is a point mutation that removes a stop codon. About 10% of patients fa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Missense Mutations
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution. Substitution of protein from DNA mutations Missense mutation refers to a change in one amino acid in a protein, arising from a point mutation in a single nucleotide. Missense mutation is a type of nonsynonymous substitution in a DNA sequence. Two other types of nonsynonymous substitution are the nonsense mutations, in which a codon is changed to a premature stop codon that results in truncation of the resulting protein, and the nonstop mutations, in which a stop codon erasement results in a longer, nonfunctional protein. Missense mutations can render the resulting protein nonfunctional, and such mutations are responsible for human diseases such as Epidermolysis bullosa, sickle-cell disease, SOD1 mediated ALS, and a substantial number of cancers. In the most common variant of sickle-cell ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autosomal Dominant
In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new (''de novo'') or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child (see Sex linkage). Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance such as incomple ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |