|
Animal Heme-dependent Peroxidases
Animal heme-dependent peroxidases is a family of peroxidases. Peroxidases are found in bacteria, fungi, plants and animals. On the basis of sequence similarity, a number of animal heme peroxidases can be categorized as members of a superfamily: myeloperoxidase (MPO); eosinophil peroxidase (EPO); lactoperoxidase (LPO); thyroid peroxidase (TPO); prostaglandin H synthase (PGHS); and peroxidasin. Function Myeloperoxidase (MPO) plays a major role in the oxygen-dependent microbicidal system of neutrophils. EPO from eosinophilic granulocytes participates in immunological reactions, and potentiates tumor necrosis factor (TNF) production and hydrogen peroxide release by human monocyte-derived macrophages. MPO (and possibly EPO) primarily use Cl−ions and H2O2 to form hypochlorous acid (HOCl), which can effectively kill bacteria or parasites. In secreted fluids, LPO catalyses the oxidation of thiocyanate ions (SCN−) by H2O2, producing the weak oxidizing agent hypothiocyanite (OSCN−), wh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Myeloperoxidase
Myeloperoxidase (MPO) is a peroxidase enzyme that in humans is encoded by the ''MPO'' gene on chromosome 17. MPO is most abundantly expressed in neutrophils (a subtype of white blood cells), and produces hypohalous acids to carry out their antimicrobial activity, including hypochlorous acid, the sodium salt of which is the chemical in bleach. It is a lysosomal protein stored in azurophilic granules of the neutrophil and released into the extracellular space during degranulation. Neutrophil myeloperoxidase has a heme pigment, which causes its green color in secretions rich in neutrophils, such as mucus and sputum. The green color contributed to its outdated name verdoperoxidase. Myeloperoxidase is found in many different organisms including mammals, birds, fish, reptiles, and amphibians. Myeloperoxidase deficiency is a well-documented disease among humans resulting in impaired immune function. Function MPO is a member of the XPO subfamily of peroxidases and produces hypoch ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eosinophil Peroxidase
Eosinophil peroxidase is an enzyme found within the eosinophil granulocytes, innate immune cells of humans and mammals. This oxidoreductase protein is encoded by the gene ''EPX'', expressed within these myeloid cells. EPO shares many similarities with its orthologous peroxidases, myeloperoxidase (MPO), lactoperoxidase (LPO), and thyroid peroxidase (TPO). The protein is concentrated in secretory granule (cell biology), granules within eosinophils. Eosinophil peroxidase is a heme peroxidase, its activities including the oxidation of halide ions to bacteriocidal reactive oxygen species, the cationic disruption of bacterial cell walls, and the post-translational modification of protein amino acid residues. The major function of eosinophil peroxidase is to catalysis, catalyze the formation of hypohalous acids from hydrogen peroxide and halide ions in solution. For example: : H2O2 + bromide, Br− → hypobromous acid, HOBr + water, H2O Hypohalous acids formed from halides or pseudohal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Families
A protein family is a group of evolutionarily related proteins. In many cases, a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term "protein family" should not be confused with family as it is used in taxonomy. Proteins in a family descend from a common ancestor and typically have similar three-dimensional structures, functions, and significant sequence similarity. Sequence similarity (usually amino-acid sequence) is one of the most common indicators of homology, or common evolutionary ancestry. Some frameworks for evaluating the significance of similarity between sequences use sequence alignment methods. Proteins that do not share a common ancestor are unlikely to show statistically significant sequence similarity, making sequence alignment a powerful tool for identifying the members of protein families. Families are sometimes grouped together into larger clades called superfamilies based on stru ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domains
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer after ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Swiss Institute Of Bioinformatics
The SIB Swiss Institute of Bioinformatics is an academic not-for-profit foundation which federates bioinformatics activities throughout Switzerland. The institute was established on 30 March 1998 and its mission is to provide core bioinformatics resources to the national and international life science research community in fields such as genomics, proteomics and systems biology as well as to lead and coordinate the field of bioinformatics in Switzerland. In particular, it promotes research, develops databanks and computer technologies, and is involved in teaching and service activities. History The institute was originally created to provide a framework for stable long-term funding for both the Swiss-Prot database and the Swiss EMBnet node. Swiss-Prot in particular went through a major funding crisis in 1996, which led the leaders of the five research groups active in bioinformatics in Geneva and Lausanne, Ron Appel, Amos Bairoch, Philipp Bucher, Victor Jongeneel and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
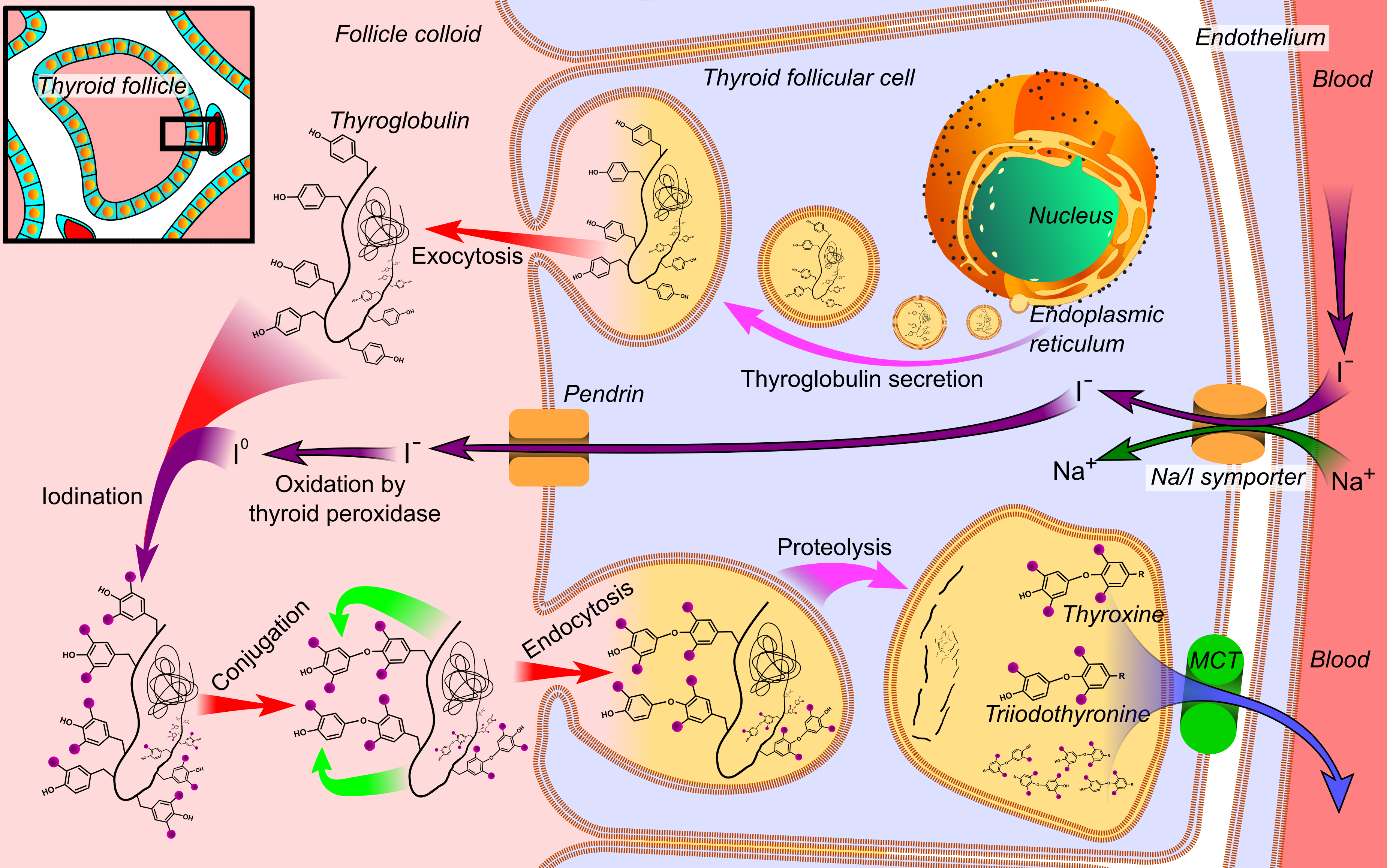
Thyroid Peroxidase
Thyroid peroxidase, also called thyroperoxidase (TPO), thyroid specific peroxidase or iodide peroxidase, is an enzyme expressed mainly in the thyroid where it is secreted into colloid. Thyroid peroxidase oxidizes iodide ions to form iodine atoms for addition onto tyrosine residues on thyroglobulin for the production of thyroxine (T4) or triiodothyronine (T3), the thyroid hormones. In humans, thyroperoxidase is encoded by the ''TPO'' gene. Function Inorganic iodine enters the body primarily as iodide, I−. After entering the thyroid follicle (or thyroid follicular cell) via a Na+/I− symporter (NIS) on the basolateral side, iodide is shuttled across the apical membrane into the colloid via pendrin after which thyroid peroxidase oxidizes iodide to atomic iodine (I) or iodinium (I+). The chemical reactions catalyzed by thyroid peroxidase occur on the outer apical membrane surface and are mediated by hydrogen peroxide. The "organification of iodine", the incorporation of iodine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PTGS2
Cyclooxygenase-2 (COX-2), also known as prostaglandin-endoperoxide synthase 2 ( HUGO PTGS2), is an enzyme that in humans is encoded by the ''PTGS2'' gene. In humans it is one of three cyclooxygenases. It is involved in the conversion of arachidonic acid to prostaglandin H2, an important precursor of prostacyclin, which is expressed in inflammation. Function PTGS2 (COX-2), converts arachidonic acid (AA) to prostaglandin endoperoxide H2. PTGSs are targets for NSAIDs and PTGS2 (COX-2) specific inhibitors called coxibs. PTGS-2 is a sequence homodimer. Each monomer of the enzyme has a peroxidase and a PTGS (COX) active site. The PTGS (COX) enzymes catalyze the conversion of AA to prostaglandins in two steps. First, hydrogen is abstracted from carbon 13 of arachidonic acid, and then two molecules of oxygen are added by the PTGS2 (COX-2), giving PGG2. Second, PGG2 is reduced to PGH2 in the peroxidase active site. The synthesized PGH2 is converted to prostaglandins ( PGD2, PG ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PTGS1
Cyclooxygenase 1 (COX-1), also known as prostaglandin-endoperoxide synthase 1 ( HUGO PTGS1), is an enzyme that in humans is encoded by the ''PTGS1'' gene. In humans it is one of three cyclooxygenases. History Cyclooxygenase (COX) is the central enzyme in the biosynthetic pathway to prostaglandins from arachidonic acid. This protein was isolated more than 40 years ago and cloned in 1988. Gene and isozymes There are two isozymes of COX encoded by distinct gene products: a constitutive COX-1 (this enzyme) and an inducible COX-2, which differ in their regulation of expression and tissue distribution. The expression of these two transcripts is differentially regulated by relevant cytokines and growth factors. This gene encodes COX-1, which regulates angiogenesis in endothelial cells. COX-1 is also involved in cell signaling and maintaining tissue homeostasis. A splice variant of COX-1 termed COX-3 was identified in the central nervous system of dogs, but does not result in a f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
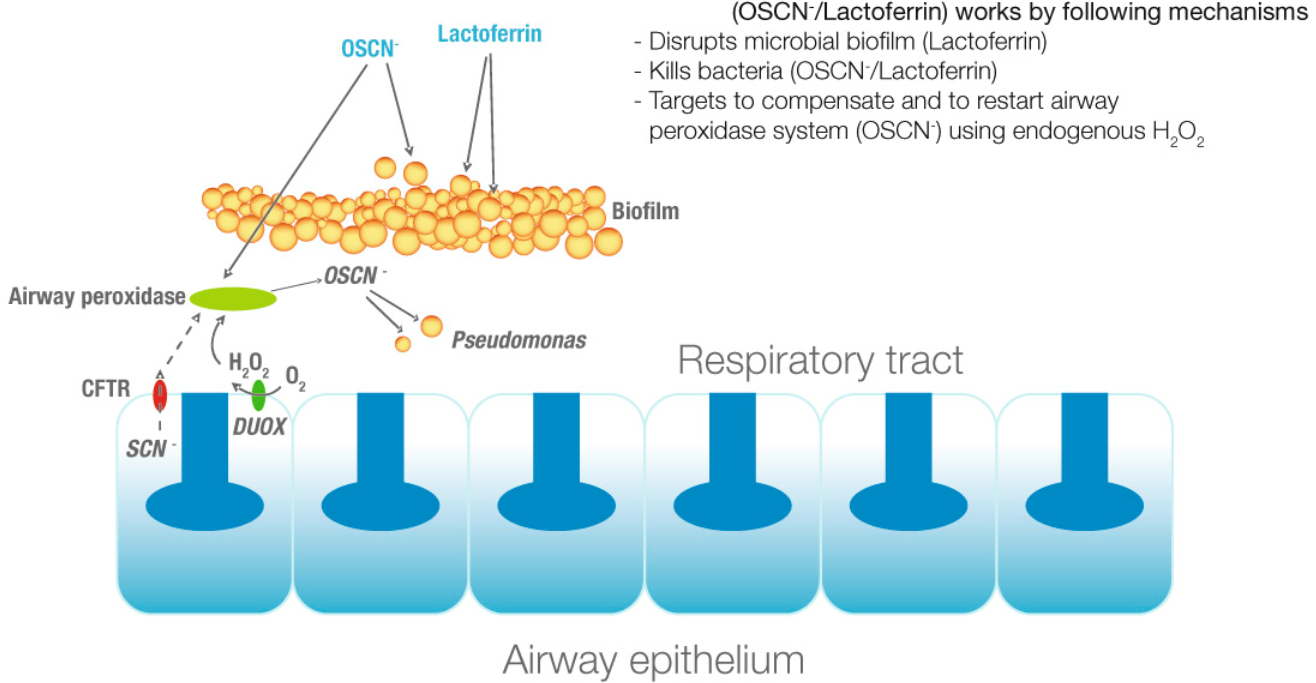
LPO (gene)
Lactoperoxidase (LPO, ) is a peroxidase enzyme secreted from mammary, salivary, tears and other mucosal glands including the lungs, bronchii and nose that function as a natural, first line of defense against bacteria and viral agents. Lactoperoxidase is a member of the heme peroxidase family of enzymes. In humans, lactoperoxidase is encoded by the ''LPO'' gene. Lactoperoxidase catalyzes the oxidation of several inorganic and many organic substrates by hydrogen peroxide. Lactoperoxidase rapidly oxidizes iodide and slowly oxidizes bromide and is designated a haloperoxidase. Another important substrate is the pseudo-halide thiocyanate. The oxidized products display potent, non-specific bactericidal and antiviral activities, including destruction of the influenza virus. Lactoperoxidase together with its inorganic ion substrates, hydrogen peroxide, DUOX1 and DUOX2 and products are termed the lactoperoxidase system. Hence LPO is considered a very important defense against invasive ba ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DUOX2
Dual oxidase 2, also known as DUOX2 or ThOX2 (for thyroid oxidase), is an enzyme that in humans is encoded by the ''DUOX2'' gene. Dual oxidase is an enzyme that was first identified in the mammalian thyroid gland. In humans, two isoforms are found; hDUOX1 and hDUOX2 (this enzyme). The protein location is not exclusive to thyroid tissue; hDUOX1 is prominent in airway epithelial cells and hDUOX2 in the salivary glands and gastrointestinal tract. Function Investigations into reactive oxygen species ( ROS) in biological systems have, until recently, focused on characterization of phagocytic cell processes. It is now well accepted that production of such species is not restricted to phagocytic cells and can occur in eukaryotic non-phagocytic cell types via NADPH oxidase (NOX) or dual oxidase (DUOX). This new family of proteins, termed the NOX/DUOX family or NOX family of NADPH oxidases, consists of homologs to the catalytic moiety of phagocytic NADPH-oxidase, gp91phox. Members of t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Peroxidase
Peroxidases or peroxide reductases ( EC numberbr>1.11.1.x are a large group of enzymes which play a role in various biological processes. They are named after the fact that they commonly break up peroxides, and should not be confused with other enzymes that ''produce'' peroxide, which are often oxidases. Functionality Peroxidases typically catalyze a reaction of the form: :ROOR' + \overset + 2H+ -> ce + R'OH Optimal substrates For many of these enzymes the optimal substrate is hydrogen peroxide, but others are more active with organic hydroperoxides such as lipid peroxides. Peroxidases can contain a heme cofactor in their active sites, or alternately redox-active cysteine or selenocysteine residues. The nature of the electron donor is very dependent on the structure of the enzyme. * For example, horseradish peroxidase can use a variety of organic compounds as electron donors and acceptors. Horseradish peroxidase has an accessible active site, and many compounds can re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |