|
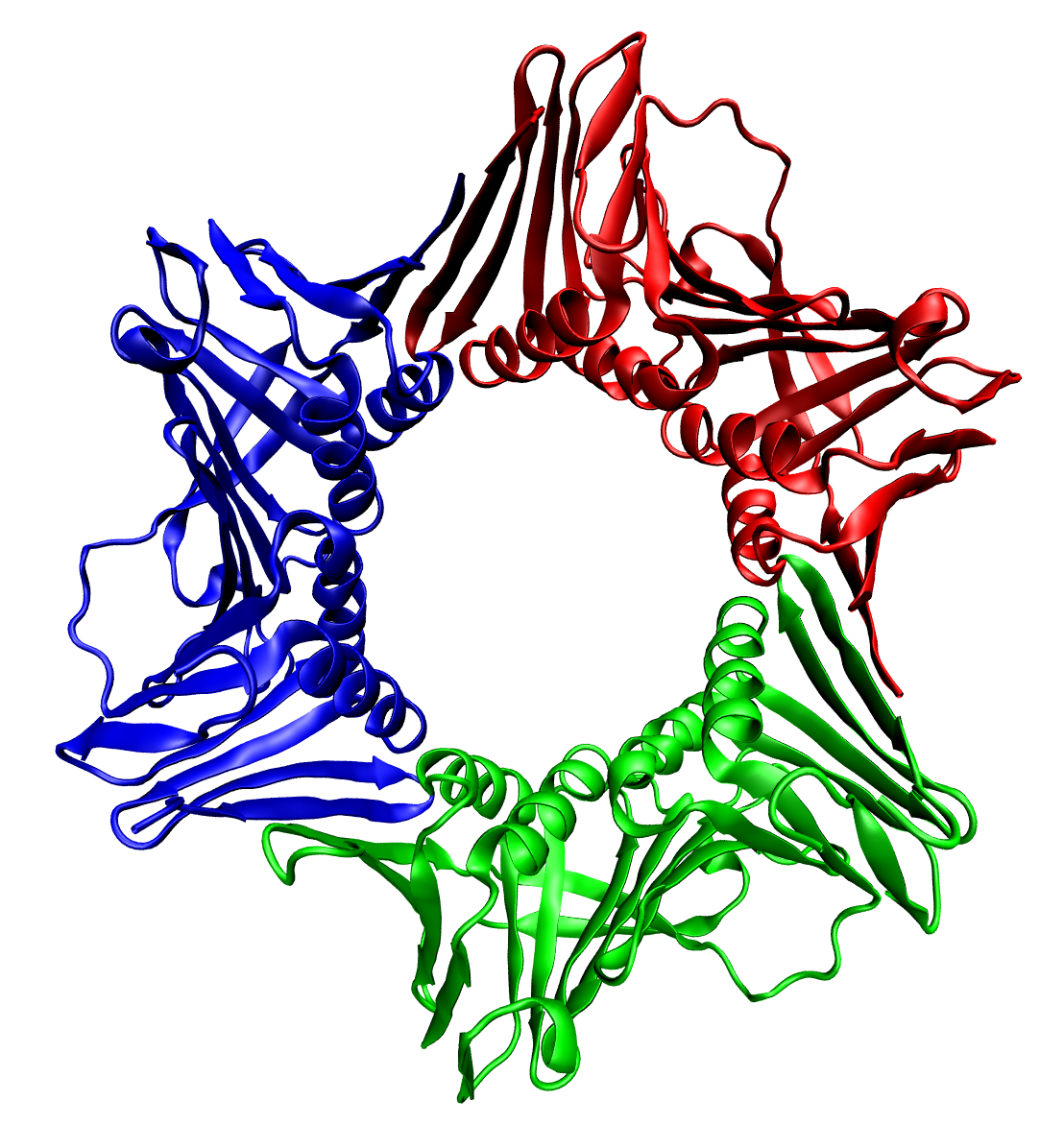
YgbB N Terminal Protein Domain
In molecular biology, YgbB is a protein domain. This entry makes reference to a number of proteins from eukaryotes and prokaryotes which share this common N-terminal signature and appear to be involved in terpenoid biosynthesis. The YgbB protein is a putative enzyme thought to aid terpenoid and isoprenoid biosynthesis, a vital chemical in all living organisms. This protein domain is part of an enzyme which catalyses a reaction in a complex pathway. Function The YgbB protein domain has a main function of being involved in terpenoid and isoprenoid biosynthesis. Biochemistry MECDP (2-C-methyl-D-erythritol 2,4-cyclodiphosphate) synthetase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis, isoprenoids being essential in all organisms. Isoprenoids can also be synthesized through the mevalonate pathway. The non-mevolante route is used by many bacteria and human pathogens, including ''Mycobacterium tuberculosis'' and ''Plasmodium falciparum''. This route appears to involve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or Disulfide bond, disulfide bridges. Domains often form functional units, such as the calcium-binding EF-hand, EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera (protein), chimeric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmodium
''Plasmodium'' is a genus of unicellular eukaryotes that are obligate parasites of vertebrates and insects. The life cycles of ''Plasmodium'' species involve development in a Hematophagy, blood-feeding insect host (biology), host which then injects parasites into a vertebrate host during a blood meal. Parasites grow within a vertebrate body tissue (often the liver) before entering the bloodstream to infect red blood cells. The ensuing destruction of host red blood cells can result in malaria. During this infection, some parasites are picked up by a blood-feeding insect (mosquitoes in majority cases), continuing the life cycle. ''Plasmodium'' is a member of the phylum Apicomplexa, a large group of parasitic eukaryotes. Within Apicomplexa, ''Plasmodium'' is in the order Haemosporida and family Plasmodiidae. Over 200 species of ''Plasmodium'' have been described, many of which have been subdivided into 14 subgenera based on parasite morphology and host range. Evolutionary relations ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding site'', and residues that catalyse a reaction of that substrate, the ''catalytic site''. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their functio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Trimer (biochemistry)
thumbnail, 400px, Trimeric form of a TNF-α mutant In biochemistry, a protein trimer is a macromolecular complex formed by three, usually non-covalently bound, macromolecules like proteins or nucleic acids. A protein trimer often occurs from the assembly of a protein's quaternary structure. The non-covalent interactions between the hydrophobic and hydrophilic regions on the polypeptides units help to stabilize the quaternary structure. Since a protein trimer is composed of multiple polypeptide subunits, it is considered an oligomer. A homotrimer would be formed by three identical molecules. A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein, while Type I collagen is an AAB-type heterotrimeric protein. An example of viral protein homotrimeric protein is mammarenavirus of Z matrix protein. Porins usually arrange themselves in membranes as trimers. __TOC__ Bacteriophage T4 tail fiber Multiple copies of a poly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Product (chemistry)
Products are the species formed from chemical reactions. During a chemical reaction, reactants are transformed into products after passing through a high energy transition state. This process results in the consumption of the reactants. It can be a spontaneous reaction or mediated by catalysts which lower the energy of the transition state, and by solvents which provide the chemical environment necessary for the reaction to take place. When represented in chemical equations, products are by convention drawn on the right-hand side, even in the case of reversible reactions. The properties of products such as their energies help determine several characteristics of a chemical reaction, such as whether the reaction is exergonic or endergonic. Additionally, the properties of a product can make it easier to extract and purify following a chemical reaction, especially if the product has a different state of matter than the reactants. Spontaneous reaction : R \rightarrow P *W ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Diphosphate
In chemistry, pyrophosphates are phosphorus oxyanions that contain two phosphorus atoms in a linkage. A number of pyrophosphate salts exist, such as disodium pyrophosphate () and tetrasodium pyrophosphate (), among others. Often pyrophosphates are called diphosphates. The parent pyrophosphates are derived from partial or complete neutralization of pyrophosphoric acid. The pyrophosphate bond is also sometimes referred to as a phosphoanhydride bond, a naming convention which emphasizes the loss of water that occurs when two phosphates form a new bond, and which mirrors the nomenclature for anhydrides of carboxylic acids. Pyrophosphates are found in ATP and other nucleotide triphosphates, which are important in biochemistry. The term pyrophosphate is also the name of esters formed by the condensation of a phosphorylated biological compound with inorganic phosphate, as for dimethylallyl pyrophosphate. This bond is also referred to as a high-energy phosphate bond. Acidity Pyr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
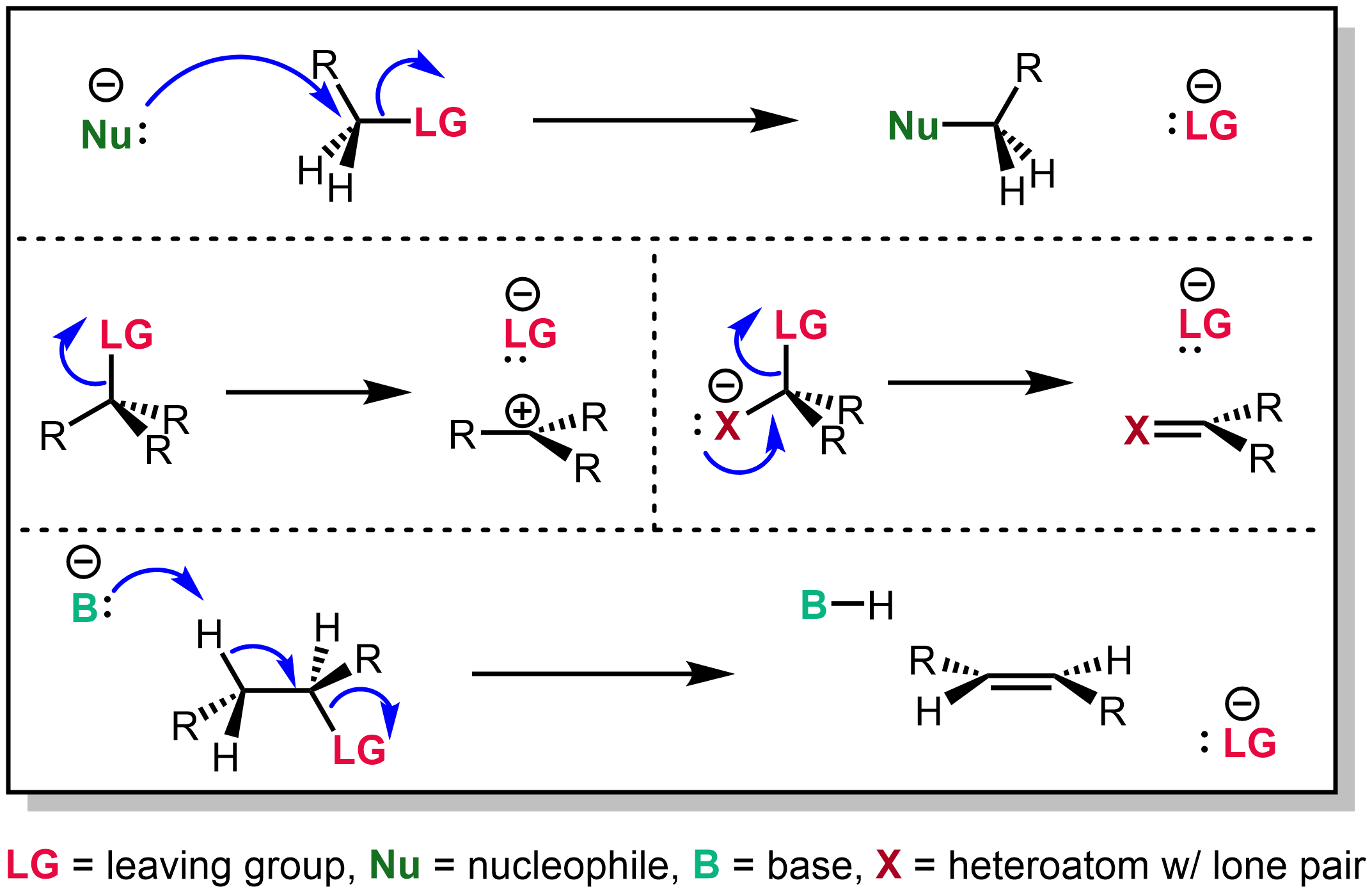
Leaving Group
In organic chemistry, a leaving group typically means a Chemical species, molecular fragment that departs with an electron, electron pair during a reaction step with heterolysis (chemistry), heterolytic bond cleavage. In this usage, a ''leaving group'' is a less formal but more commonly used synonym of the term ''nucleofuge''; although IUPAC gives the term a broader definition. A species' ability to serve as a leaving group can affect whether a reaction is viable, as well as what mechanism the reaction takes. Leaving group ability depends strongly on context, but often correlates with ability to stabilize additional electron density from bond heterolysis. Common anionic leaving groups are , and halides and sulfonate esters such as tosylate (). Water (), alcohols (), and amines () are common neutral leaving groups, although they often require activating catalysts. Some moieties, such as hydride (H−) serve as leaving groups only extremely rarely. Nomenclature IUPAC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytidine Monophosphate
Cytidine monophosphate, also known as 5'-cytidylic acid or simply cytidylate, and abbreviated CMP, is a nucleotide that is used as a monomer in RNA. It is an ester of phosphoric acid with the nucleoside cytidine. CMP consists of the phosphate group, the pentose sugar ribose, and the nucleobase cytosine; hence, a ribonucleoside monophosphate. As a substituent it takes the form of the prefix cytidylyl-. Metabolism CMP can be phosphorylated to cytidine diphosphate by the enzyme CMP kinase, with adenosine triphosphate or guanosine triphosphate donating the phosphate group. Since cytidine triphosphate is generated by amination of uridine triphosphate, the main source of CMP is from RNA being decomposed by RNAse. See also * Deoxycytidine monophosphate * Nucleoside * Oligonucleotide Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the labor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphate
Phosphates are the naturally occurring form of the element phosphorus. In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosphoric acid, phosphoric acid . The phosphate or orthophosphate ion is derived from phosphoric acid by the removal of three protons . Removal of one proton gives the dihydrogen phosphate ion while removal of two protons gives the hydrogen phosphate ion . These names are also used for salts of those anions, such as ammonium dihydrogen phosphate and trisodium phosphate. File:3-phosphoric-acid-3D-balls.png, Phosphoricacid File:2-dihydrogenphosphate-3D-balls.png, Dihydrogenphosphate File:1-hydrogenphosphate-3D-balls.png, Hydrogenphosphate File:0-phosphate-3D-balls.png, Phosphate or orthophosphate In organic chemistry, phosphate or orthophosphate is an organophosphate, an ester of orthophosphoric acid of the form where one ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Catalysis
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quickly, very small amounts of catalyst often suffice; mixing, surface area, and temperature are important factors in reaction rate. Catalysts generally react with one or more reactants to form intermediates that subsequently give the final reaction product, in the process of regenerating the catalyst. The rate increase occurs because the catalyst allows the reaction to occur by an alternative mechanism which may be much faster than the noncatalyzed mechanism. However the noncatalyzed mechanism does remain possible, so that the total rate (catalyzed plus noncatalyzed) can only increase in the presence of the catalyst and never decrease. Catalysis may be classified as either homogeneous, whose components are dispersed in the same phase (usual ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synthetase
In biochemistry, a ligase is an enzyme that can catalyze the joining ( ligation) of two molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the molecules, typically resulting in the formation of new C-O, C-S, or C-N bonds. For example, DNA ligase can join two complementary fragments of nucleic acid by forming phosphodiester bonds, and repair single stranded breaks that arise in double stranded DNA during replication. In general, a ligase catalyzes the following dehydration reaction, thus joining molecules A and B: A-OH + B-H → A–B + H2O Nomenclature The naming of ligases is inconsistent and so these enzymes are commonly known by several different names. Generally, the common names of ligases include the word "ligase", such as in DNA ligase, an enzyme commonly used in molecular biology laboratories to join together DNA fragments. However, many common names use the term "synthetase" or "synthase" instead, bec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymes
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include catalytic RNA molecules, also called ribozymes. They are sometimes described as a ''type'' of enzyme rather than being ''like'' an enzyme, but even in the d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |