|
SULF1
Sulfatase 1, also known as SULF1, is an enzyme which in humans is encoded by the ''SULF1'' gene. Heparan sulfate proteoglycans ( HSPGs) act as co-receptors for numerous heparin-binding growth factors and cytokines and are involved in cell signaling. Heparan sulfate 6-O-endo-sulfatases, such as SULF1, selectively remove 6-O-sulfate groups from heparan sulfate. This activity modulates the effects of heparan sulfate by altering binding sites for signaling molecules. Function Heparan sulfate proteoglycans (HSPGs) are widely expressed throughout most tissues of nearly all multicellular species. The function of HSPGs extends beyond providing an extracellular matrix (ECM) structure and scaffold for cells. They are integral regulators of essential cell signaling pathways affecting cell growth, proliferation, differentiation, and migration. Although the core protein is important, the large heparan sulfate (HS) chains extending from the core are responsible for most receptor signalin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
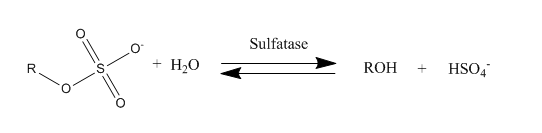
Sulfatase
Sulfatases are enzymes of the esterase class that catalyze the hydrolysis of sulfate esters. These may be found on a range of substrates, including steroids, carbohydrates and proteins. Sulfate esters may be formed from various alcohols and amines. In the latter case the resultant N-sulfates can also be termed sulfamates. Sulfatases play important roles in the cycling of sulfur in the environment, in the degradation of sulfated glycosaminoglycans and glycolipids in the lysosome, and in remodelling sulfated glycosaminoglycans in the extracellular space. Together with sulfotransferases, sulfatases form the major catalytic machinery for the synthesis and breakage of sulfate esters. Occurrence and importance Sulfatases are found in lower and higher organisms. In higher organisms they are found in intracellular and extracellular spaces. Steroid sulfatase is distributed in a wide range of tissues throughout the body, enabling sulfated steroids synthesized in the adrenals and gona ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fibroblast Growth Factor
Fibroblast growth factors (FGF) are a family of cell signalling proteins produced by macrophages; they are involved in a wide variety of processes, most notably as crucial elements for normal development in animal cells. Any irregularities in their function lead to a range of developmental defects. These growth factors typically act as systemic or locally circulating molecules of extracellular origin that activate cell surface receptors. A defining property of FGFs is that they bind to heparin and to heparan sulfate. Thus, some are sequestered in the extracellular matrix of tissues that contains heparan sulfate proteoglycans and are released locally upon injury or tissue remodeling. Families In humans, 23 members of the FGF family have been identified, all of which are ''structurally'' related signaling molecules: * Members FGF1 through FGF10 all bind fibroblast growth factor receptors (FGFRs). FGF1 is also known as ''acidic fibroblast growth factor'', and FGF2 is also kno ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PANC-1
PANC-1 is a human pancreatic cancer cell line isolated from a pancreatic carcinoma of Ductal cells, ductal cell origin. PANC-1 was derived from the tissue of a 56-year-old male. The cells can metastasis, metastasize but have poor differentiation abilities. PANC-1 cells take 52 hours to double in population, have a modal chromosome number of 63, and show G6PD of the slow mobility type. PANC-1 cells are known to have an epithelial morphology and are adherent in cell culture flasks. The cells can be frozen and regrown in culture, provided that they are appropriately warmed. Additionally, PANC-1 cells have a tendency to clump, a feature which can be avoided with trypsinization. PANC-1 cells have been used to study the role of keratin reorganization during the migration of cancer cells, along with calcium-mediated actin reset in response to physiological changes. See also *DU145 *BxPC-3 *MIA PaCa-2 References {{Reflist External linksCellosaurus entry for PANC-1 Human cell lines ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Deacetylase
Histone deacetylases (, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly. This is important because DNA is wrapped around histones, and DNA expression is regulated by acetylation and de-acetylation. Its action is opposite to that of histone acetyltransferase. HDAC proteins are now also called lysine deacetylases (KDAC), to describe their function rather than their target, which also includes non-histone proteins. HDAC super family Together with the acetylpolyamine amidohydrolases and the acetoin utilization proteins, the histone deacetylases form an ancient protein superfamily known as the histone deacetylase superfamily. Classes of HDACs in higher eukaryotes HDACs, are classified in four classes depending on sequence homology to the yeast original enzymes and domain organization: HDAC (except class III) contain zinc and are known as Zn2+-dependen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Human Umbilical Vein Endothelial Cell
Human umbilical vein endothelial cells (HUVECs) are cells derived from the endothelium of veins from the umbilical cord. They are used as a laboratory model system for the study of the function and pathology of endothelial cells (e.g., angiogenesis). They are used due to their low cost, and simple techniques for isolating them from umbilical cords, which are normally resected after childbirth. HUVECs were first isolated and cultured ''in vitro'' in the 1970s by Jaffe and others. Jiménez, N., Krouwer, V. & Post, J. A new, rapid and reproducible method to obtain high quality endothelium in vitro. Cytotechnology 65, 1-14 (2012). HUVECs can be easily made to proliferate in a laboratory setting. Like human umbilical artery endothelial cells they exhibit a cobblestone phenotype when lining vessel walls. Inhibition of the sirtuin protein sirtuin 1 (SIRT1) in HUVECs has been shown to induce cellular senescence. Conversely, overexpression of SIRT1 in HUVECs has been shown to inhibit ce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-acetylglucosamine-6-sulfatase
''N''-acetylglucosamine-6-sulfatase (EC 3.1.6.14, glucosamine (''N''-acetyl)-6-sulfatase, systematic name ''N''-acetyl-D-glucosamine-6-sulfate 6-sulfohydrolase) is an enzyme that in humans is encoded by the GNS gene. It is deficient in Sanfilippo Syndrome type IIId. It catalyses the hydrolysis of the 6-sulfate groups of the ''N''-acetyl-D-glucosamine 6-sulfate units of heparan sulfate and keratan sulfate Function ''N''-acetylglucosamine-6-sulfatase is a lysosomal enzyme found in all cells. It is involved in the catabolism of heparin, heparan sulphate, and keratan sulphate. Clinical significance Deficiency of this enzyme results in the accumulation of undergraded substrate and the lysosomal storage disorder mucopolysaccharidosis type IIID (Sanfilippo D syndrome). Mucopolysaccharidosis type IIID is the least common of the four subtypes of Sanfilippo syndrome. Nomenclature The systematic name A systematic name is a name given in a systematic way to one unique group, organi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arylsulfatase B
Arylsulfatase B (N-acetylgalactosamine-4-sulfatase, chondroitinsulfatase, chondroitinase, acetylgalactosamine 4-sulfatase, N-acetylgalactosamine 4-sulfate sulfohydrolase, ) is an enzyme associated with mucopolysaccharidosis VI (Maroteaux–Lamy syndrome). Arylsulfatase B is among a group of arylsulfatase enzymes present in the lysosomes of the liver, pancreas, and kidneys of animals. The purpose of the enzyme is to hydrolyze sulfates in the body. ARSB does this by breaking down glycosaminoglycans (GAGs), which are large sugar molecules in the body. ARSB targets two GAGs in particular: dermatan sulfate and chondroitin sulfate.U.S. National Library of Medicine"ARSB" Genetics Home Resource, 7 November 2010, Retrieved 22 November 2010 Over 130 mutations to ARSB have been found, each leading to a deficiency in the body. In most cases, the mutation occurs on a single nucleotide in the sequence. An arylsulfatase B deficiency can lead to an accumulation of GAGs in lysosomes, which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arylsulfatase A
Arylsulfatase A (or cerebroside-sulfatase) is an enzyme that breaks down sulfatides, namely cerebroside 3-sulfate into cerebroside and sulfate. In humans, arylsulfatase A is encoded by the ''ARSA'' gene. Pathology A deficiency is associated with metachromatic leukodystrophy, an autosomal recessive In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and ... disease. Biochemistry Enzyme regulation Arylsulfatase A is inhibited by phosphate, which forms a covalent bond with the active site 3-oxoalanine. ; References Further reading * * * * * * * * * * * * * * * * * * External links GeneReviews/NCBI/NIH/UW entry on Arylsulfatase A Deficiency - Metachromatic Leukodystrophy OMIM entries on ARSA Deficiency * * * {{hydrolase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arylsulfatase
Arylsulfatase (EC 3.1.6.1, sulfatase, nitrocatechol sulfatase, phenolsulfatase, phenylsulfatase, ''p''-nitrophenyl sulfatase, arylsulfohydrolase, 4-methylumbelliferyl sulfatase, estrogen sulfatase) is a type of sulfatase enzyme with systematic name ''aryl-sulfate sulfohydrolase''. This enzyme catalyses the following chemical reaction : an aryl sulfate + H2O \rightleftharpoons a phenol + sulfate Types include: *Arylsulfatase A (also known as "cerebroside-sulfatase") *Arylsulfatase B (also known as "''N''-acetylgalactosamine-4-sulfatase") *Steroid sulfatase (formerly known as "arylsulfatase C") * ARSC2 * ARSD *ARSE * ARSF * ARSG * ARSH * ARSI * ARSJ *ARSK See also * Aryl In organic chemistry, an aryl is any functional group or substituent derived from an aromatic ring, usually an aromatic hydrocarbon, such as phenyl and naphthyl. "Aryl" is used for the sake of abbreviation or generalization, and "Ar" is used as ... References Hydrolases EC 3.1.6 {{biochem-s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Frizzled
Frizzled is a family of atypical G protein-coupled receptors that serve as receptors in the Wnt signaling pathway and other signaling pathways. When activated, Frizzled leads to activation of Dishevelled in the cytosol. Species distribution Frizzled proteins and the genes that encode them have been identified in an array of animals, from sponges to humans. Function Frizzled proteins also play key roles in governing cell polarity, embryonic development, formation of neural synapses, cell proliferation, and many other processes in developing and adult organisms. These processes occur as a result of one of three signaling pathways. These include the canonical Wnt/β-catenin pathway, Wnt/calcium pathway, and planar cell polarity (PCP) pathway. Mutations in the human frizzled-4 receptor have been linked to familial exudative vitreoretinopathy, a rare disease affecting the retina at the back of the eye, and the vitreous, the clear fluid inside the eye. The frizzled (fz) lo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |