|
Molecular Beacon
Molecular beacons, or molecular beacon probes, are oligonucleotide hybridization probes that can report the presence of specific nucleic acids in homogenous solutions. Molecular beacons are Stem-loop, hairpin-shaped molecules with an internally Quenching (fluorescence), quenched fluorophore whose fluorescence is restored when they bind to a target nucleic acid sequence. This is a novel non-radioactive method for detecting specific sequences of nucleic acids. They are useful in situations where it is either not possible or desirable to isolate the probe-target hybrids from an excess of the hybridization probes. Molecular beacon probes A typical molecular beacon probe is 25 nucleotides long. The middle 15 nucleotides are complementary to the target DNA or RNA and do not base pair with one another, while the five nucleotides at each terminus are complementary to each other rather than to the target DNA. A typical molecular beacon structure can be divided in 4 parts: 1) loop, an 18–3 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase chemical synthesis, these small fragments of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as Fluorescence in situ hybridization, molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the Nucleic acid sequence, sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by "Monomer, -m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
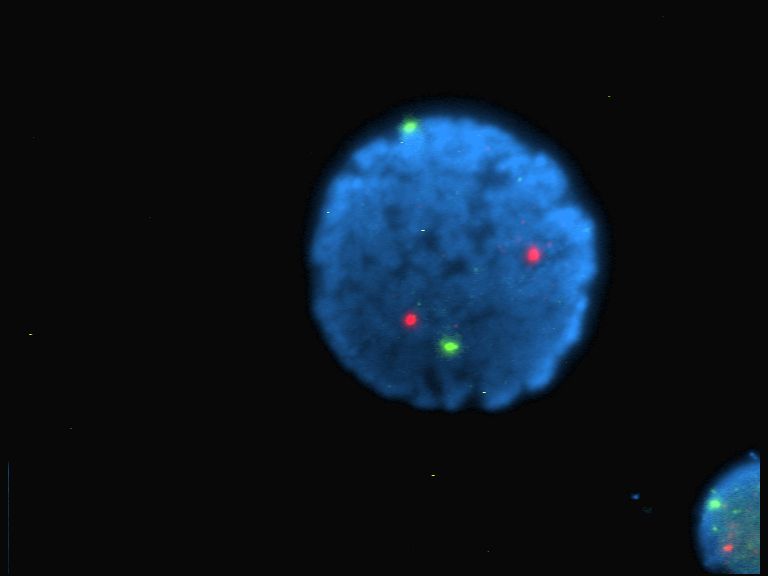
Hybridization Probe
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA, usually 15–10000 nucleotides long, which can be radioactively or fluorescently labeled. HPs can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured (by heating or under alkaline conditions such as exposure to sodium hydroxide) into single stranded DNA (ssDNA) and then hybridized to the target ssDNA ( Southern blotting) or RNA ( northern blotting) immobilized on a membrane or '' in situ''. To detect hybridization of the probe to its target sequence, the probe is tagged (or "labeled") with a molecular marker of either radioactive or (more recently) fluorescent molecules. Commonly used markers are 32P (a radioactive isotope of phosphorus incorporated into the phosphodiester bond in the probe DNA), digoxigenin, a non-radioactive, antibody-based marker, biotin or fluorescein. DNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a pentose, 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is deoxyribose, a variant of ribose, the polymer is DNA. Nucleic acids are chemical compounds that are found in nature. They carry information in cells and make up genetic material. These acids are very common in all living things, where they create, encode, and store information in every living cell of every outline of life forms, life-form on Earth. In turn, they send and express that information inside and outside the cell nucleus. From the inner workings of the cell to the young of a living thing, they contain and provide information via the nucleic acid sequence. This gives the RNA and DNA their unmistakable 'la ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regions of the same nucleic acid strand, usually Complementarity (molecular biology), complementary in nucleotide sequence, base-pair to form a double helix that ends in a loop of unpaired nucleotides. Stem-loops are most commonly found in RNA, and are a key building block of many RNA biomolecular structure#Secondary structure, secondary structures. Stem-loops can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA-binding protein, RNA binding proteins, and serve as a Substrate (chemistry), substrate for Enzyme catalysis, enzymatic reactions. Formation and stability The formation of a stem-loop is dependent on the stability of the helix and loop regions. The first prerequisite is the p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quenching (fluorescence)
In chemistry, quenching refers to any process which decreases the fluorescent intensity of a given substance. A variety of processes can result in quenching, such as excited state reactions, energy transfer, complex-formation and collisions. As a consequence, quenching is often heavily dependent on pressure and temperature. Molecular oxygen, iodine ions and acrylamide are common chemical quenchers. The chloride ion is a well known quencher for quinine fluorescence. Quenching poses a problem for non-instant spectroscopic methods, such as laser-induced fluorescence. Quenching is made use of in optode sensors; for instance the quenching effect of oxygen on certain ruthenium complexes allows the measurement of oxygen saturation in solution. Quenching is the basis for Förster resonance energy transfer (FRET) assays. Quenching and dequenching upon interaction with a specific molecular biological target is the basis for activatable optical contrast agents for molecular imaging. M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds. Fluorophores are sometimes used alone, as a tracer in fluids, as a dye for staining of certain structures, as a substrate of enzymes, or as a probe or indicator (when its fluorescence is affected by environmental aspects such as polarity or ions). More generally they are covalently bonded to macromolecules, serving as a markers (or dyes, or tags, or reporters) for affine or bioactive reagents (antibodies, peptides, nucleic acids). Fluorophores are notably used to stain tissues, cells, or materials in a variety of analytical methods, such as fluorescent imaging and spectroscopy. Fluorescein, via its amine-reactive isothiocyanate derivative fluorescein isothiocyanate (FITC), has been one of the most popular fluorophores ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence
Fluorescence is one of two kinds of photoluminescence, the emission of light by a substance that has absorbed light or other electromagnetic radiation. When exposed to ultraviolet radiation, many substances will glow (fluoresce) with colored visible light. The color of the light emitted depends on the chemical composition of the substance. Fluorescent materials generally cease to glow nearly immediately when the radiation source stops. This distinguishes them from the other type of light emission, phosphorescence. Phosphorescent materials continue to emit light for some time after the radiation stops. This difference in duration is a result of quantum spin effects. Fluorescence occurs when a photon from incoming radiation is absorbed by a molecule, exciting it to a higher energy level, followed by the emission of light as the molecule returns to a lower energy state. The emitted light may have a longer wavelength and, therefore, a lower photon energy than the absorbed radi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Radioactive
Radioactive decay (also known as nuclear decay, radioactivity, radioactive disintegration, or nuclear disintegration) is the process by which an unstable atomic nucleus loses energy by radiation. A material containing unstable nuclei is considered ''radioactive''. Three of the most common types of decay are Alpha decay, alpha, Beta decay, beta, and Gamma ray, gamma decay. The weak force is the Fundamental interactions, mechanism that is responsible for beta decay, while the other two are governed by the electromagnetic force, electromagnetic and nuclear forces. Radioactive decay is a randomness, random process at the level of single atoms. According to quantum mechanics, quantum theory, it is impossible to predict when a particular atom will decay, regardless of how long the atom has existed. However, for a significant number of identical atoms, the overall decay rate can be expressed as a decay constant or as a half-life. The half-lives of radioactive atoms have a huge range: f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Beacon Binding
A molecule is a group of two or more atoms that are held together by attractive forces known as chemical bonds; depending on context, the term may or may not include ions that satisfy this criterion. In quantum physics, organic chemistry, and biochemistry, the distinction from ions is dropped and ''molecule'' is often used when referring to polyatomic ions. A molecule may be homonuclear, that is, it consists of atoms of one chemical element, e.g. two atoms in the oxygen molecule (O2); or it may be heteronuclear, a chemical compound composed of more than one element, e.g. water (two hydrogen atoms and one oxygen atom; H2O). In the kinetic theory of gases, the term ''molecule'' is often used for any gaseous particle regardless of its composition. This relaxes the requirement that a molecule contains two or more atoms, since the noble gases are individual atoms. Atoms and complexes connected by non-covalent interactions, such as hydrogen bonds or ionic bonds, are typically not ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotides
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all Life, life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common Nutrient, nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a pentose, five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine, and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid Hybridisation
In molecular biology, hybridization (or hybridisation) is a phenomenon in which single-stranded deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) molecules anneal to complementary DNA or RNA. Though a double-stranded DNA sequence is generally stable under physiological conditions, changing these conditions in the laboratory (generally by raising the surrounding temperature) will cause the molecules to separate into single strands. These strands are complementary to each other but may also be complementary to other sequences present in their surroundings. Lowering the surrounding temperature allows the single-stranded molecules to anneal or “hybridize” to each other. DNA replication and transcription of DNA into RNA both rely upon nucleotide hybridization, as do molecular biology techniques including Southern blots and Northern blots, the polymerase chain reaction (PCR), and most approaches to DNA sequencing. Applications Hybridization is a basic property of nucleotide s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |