|
MSH6
MSH6 or mutS homolog 6 is a gene that codes for DNA mismatch repair protein Msh6 in the budding yeast ''Saccharomyces cerevisiae''. It is the homologue of the human "G/T binding protein," (GTBP) also called p160 or hMSH6 (human MSH6). The MSH6 protein is a member of the Mutator S (MutS) family of proteins that are involved in DNA damage repair. Defects in hMSH6 are associated with atypical hereditary nonpolyposis colorectal cancer not fulfilling the Amsterdam criteria for HNPCC. hMSH6 mutations have also been linked to endometrial cancer and the development of endometrial carcinomas. Discovery MSH6 was first identified in the budding yeast ''S. cerevisiae'' because of its homology to MSH2. The identification of the human GTBP gene and subsequent amino acid sequence availability showed that yeast MSH6 and human GTBP were more related to each other than any other MutS homolog, with a 26.6% amino acid identity. Thus, GTBP took on the name human MSH6, or hMSH6. Structure In the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MSH2
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the ''MSH2'' gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that codes for a DNA mismatch repair (MMR) protein, MSH2, which forms a heterodimer with MSH6 to make the human MutSα mismatch repair complex. It also dimerizes with MSH3 to form the MutSβ DNA repair complex. MSH2 is involved in many different forms of DNA repair, including transcription-coupled repair, homologous recombination, and base excision repair. Mutations in the MSH2 gene are associated with microsatellite instability and some cancers, especially with hereditary nonpolyposis colorectal cancer (HNPCC). At least 114 disease-causing mutations in this gene have been discovered. Clinical significance Hereditary nonpolyposis colorectal cancer (HNPCC), sometimes referred to as Lynch syndrome, is inherited in an autosomal dominant fashio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hereditary Nonpolyposis Colorectal Cancer
Hereditary nonpolyposis colorectal cancer (HNPCC) is a hereditary predisposition to colon cancer. HNPCC includes (and was once synonymous with) Lynch syndrome, an autosomal dominant genetic condition that is associated with a high risk of colon cancer, endometrial cancer (second most common), ovary, stomach, small intestine, hepatobiliary tract, upper urinary tract, brain, and skin. The increased risk for these cancers is due to inherited genetic mutations that impair DNA mismatch repair. It is a type of cancer syndrome. Other HNPCC conditions include Lynch-like syndrome, polymerase proofreading-associated polyposis and familial colorectal cancer type X. Signs and symptoms Risk of cancer ''Lifetime risk and mean age at diagnosis for Lynch syndrome–associated cancers'' In addition to the types of cancer found in the chart above, it is understood that Lynch syndrome also contributes to an increased risk of small bowel cancer, pancreatic cancer, ureter/renal pelvis cancer, b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PCNA
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase delta, DNA polymerase δ in eukaryotic cell (biology), cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics. Many proteins interact with PCNA via the two known PCNA-interacting motifs PCNA-interacting peptide (PIP) box and AlkB homologue 2 PCNA interacting motif (APIM). Proteins binding to PCNA via the PIP-box are mainly involved in DNA replication whereas proteins binding to PCNA via APIM are mainly important in the context of genotoxic stress. Function The protein encoded by this gene is found in the nucleus and is a cofactor of DNA polymerase delta. The encoded protein acts as a homotrimer and helps increase the processivity of leading strand synthesis during DNA replication. In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
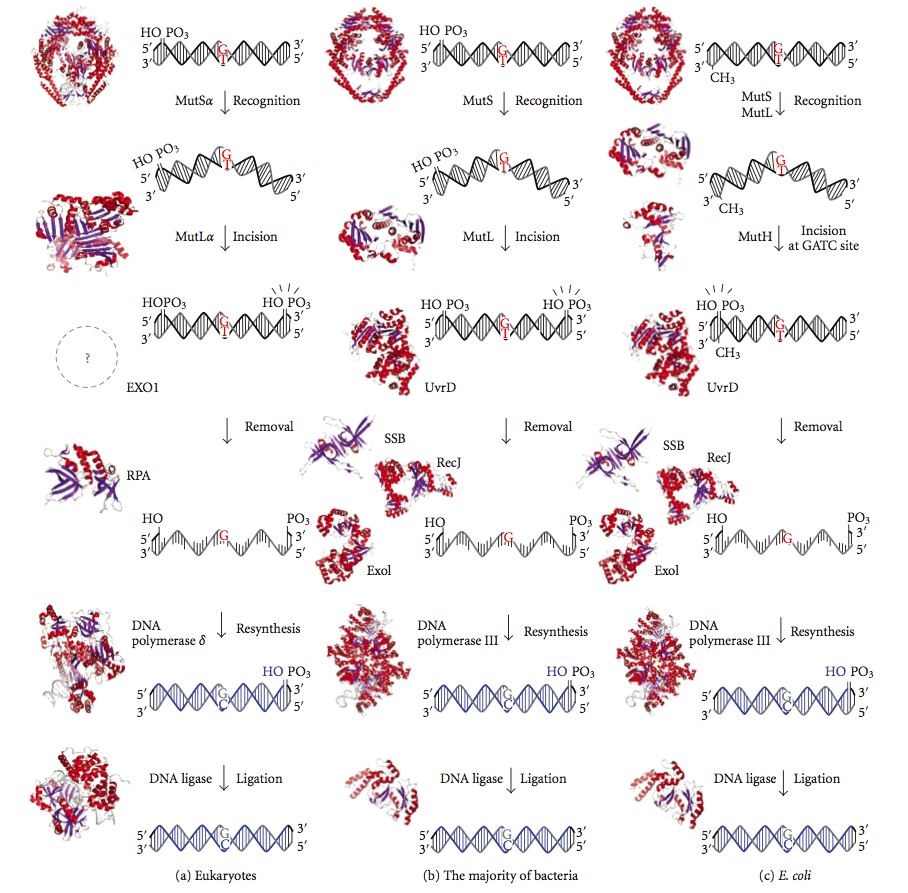
DNA Mismatch Repair
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of nucleobase, bases that can arise during DNA replication and Genetic recombination, recombination, as well as DNA repair, repairing some forms of DNA damage. Mismatch repair is strand-specific. During DNA synthesis the newly synthesised (daughter) strand will commonly include errors. In order to begin repair, the mismatch repair machinery distinguishes the newly synthesised strand from the template (parental). In gram-negative bacteria, transient methylase, hemimethylation distinguishes the strands (the parental is methylated and daughter is not). However, in other prokaryotes and eukaryotes, the exact mechanism is not clear. It is suspected that, in eukaryotes, newly synthesized lagging-strand DNA transiently contains Nick (DNA), nicks (before being sealed by DNA ligase) and provides a signal that directs mismatch proofreading systems to the appropriate st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsatellite Instability
Microsatellite instability (MSI) is the condition of genetic hypermutability (predisposition to mutation) that results from impaired DNA mismatch repair (MMR). The presence of MSI represents phenotypic evidence that MMR is not functioning normally. MMR corrects errors that spontaneously occur during DNA replication, such as single base mismatches or short insertions and deletions. The proteins involved in MMR correct polymerase errors by forming a complex that binds to the mismatched section of DNA, excises the error, and inserts the correct sequence in its place. Cells with abnormally functioning MMR are unable to correct errors that occur during DNA replication and consequently accumulate errors. This causes the creation of novel microsatellite fragments. Polymerase chain reaction-based assays can reveal these novel microsatellites and provide evidence for the presence of MSI. Microsatellites are repeated sequences of DNA. These sequences can be made of units of 1 to 6 bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a human tumor suppressor gene (also known as a caretaker gene) and is responsible for repairing DNA. ''BRCA1'' and ''BRCA2'' are unrelated proteins, but both are normally expressed in the cells of breast and other tissues, where they help repair damaged DNA, or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks. If ''BRCA1'' or ''BRCA2'' itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer. ''BRCA1'' and ''BRCA2'' have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a no ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endometrial Cancer
Endometrial cancer is a cancer that arises from the endometrium (the epithelium, lining of the uterus or womb). It is the result of the abnormal growth of cells (biology), cells that can invade or spread to other parts of the body. The first sign is most often vaginal bleeding not associated with a menstrual period. Other symptoms include dysuria, pain with urination, dyspareunia, pain during sexual intercourse, or pelvic pain. Endometrial cancer occurs most commonly after menopause. Approximately 40% of cases are related to obesity. Endometrial cancer is also associated with excessive estrogen exposure, hypertension, high blood pressure and diabetes mellitus, diabetes. Whereas taking estrogen alone increases the risk of endometrial cancer, taking both estrogen and a progestogen in combination, as in most birth control pills, decreases the risk. Between two and five percent of cases are related to genes inherited from the parents. Endometrial cancer is sometimes called "uterin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cancer Epigenetics
Cancer epigenetics is the study of epigenetics, epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than mutation, genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of gene expression, expression of genes that occurs about 10 times more frequently by transcription silencing (caused by epigenetic promoter hypermethylation of CpG site#Methylation, silencing, cancer, and aging, CpG islands) than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 Genetic hitchhiking, hitchhiker or passenger mutations. However ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CpG Site
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands. Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosines. Enzymes that add a methyl group are called DNA methyltransferases. In mammals, 70% to 80% of CpG cytosines are methylated. Methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics. Methylated cytosines often mutate to thymines. In humans, about 70% of promoters located near the transcription start site of a gene (proximal promoters) contain a CpG island. CpG characteristics Definition ''CpG'' is shorthand for ''5'—C—phosphate—G—3' '', that is, cytosine and guanine separated by only one phosphate group; phosphate links any two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MicroRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in messenger RNA (mRNA) molecules, then silence said mRNA molecules by one or more of the following processes: * Cleaving the mRNA strand into two pieces. * Destabilizing the mRNA by shortening its poly(A) tail. * Reducing translation of the mRNA into proteins. In cells of humans and other animals, miRNAs primarily act by destabilizing the mRNA. miRNAs resemble the small interfering RNAs (siRNAs) of the RNA interference (RNAi) pathway, except miRNAs derive from regions of RNA transcripts that fold back on themselves to form short stem-loops (hairpins), whereas siRNAs derive from longer regions of double-stranded RNA. The human genome may encode ov ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Overview For transcription to take place, the enzyme that synthesizes RNA, known as RNA polymerase, must attach to the DNA near a gene. Promoters contain specific DNA sequences such as response elements that provide a secure initial binding site for RNA polymerase and for proteins called transcription factors that recruit RNA polymerase. These transcription factor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |