|
MSH2
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the ''MSH2'' gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that codes for a DNA mismatch repair (MMR) protein, MSH2, which forms a heterodimer with MSH6 to make the human MutSα mismatch repair complex. It also dimerizes with MSH3 to form the MutSβ DNA repair complex. MSH2 is involved in many different forms of DNA repair, including transcription-coupled repair, homologous recombination, and base excision repair. Mutations in the MSH2 gene are associated with microsatellite instability and some cancers, especially with hereditary nonpolyposis colorectal cancer (HNPCC). At least 114 disease-causing mutations in this gene have been discovered. Clinical significance Hereditary nonpolyposis colorectal cancer (HNPCC), sometimes referred to as Lynch syndrome, is inherited in an autosomal dominant fashio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MSH6
MSH6 or mutS homolog 6 is a gene that codes for DNA mismatch repair protein Msh6 in the budding yeast ''Saccharomyces cerevisiae''. It is the homologue of the human "G/T binding protein," (GTBP) also called p160 or hMSH6 (human MSH6). The MSH6 protein is a member of the Mutator S (MutS) family of proteins that are involved in DNA damage repair. Defects in hMSH6 are associated with atypical hereditary nonpolyposis colorectal cancer not fulfilling the Amsterdam criteria for HNPCC. hMSH6 mutations have also been linked to endometrial cancer and the development of endometrial carcinomas. Discovery MSH6 was first identified in the budding yeast ''S. cerevisiae'' because of its homology to MSH2. The identification of the human GTBP gene and subsequent amino acid sequence availability showed that yeast MSH6 and human GTBP were more related to each other than any other MutS homolog, with a 26.6% amino acid identity. Thus, GTBP took on the name human MSH6, or hMSH6. Structure In the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MSH3
DNA mismatch repair protein, MutS Homolog 3 (MSH3) is a human homologue of the bacterial mismatch repair protein MutS that participates in the mismatch repair (MMR) system. MSH3 typically forms the heterodimer MutSβ with MSH2 in order to correct long insertion/deletion loops and base-base mispairs in microsatellites during DNA synthesis. Deficient capacity for MMR is found in approximately 15% of colorectal cancers, and somatic mutations in the MSH3 gene can be found in nearly 50% of MMR-deficient colorectal cancers. Gene and expression In humans, the encoding gene for MSH3 is found on chromosome 5 at location 5q11-q12 upstream of the dihydrofolate reductase (DHFR) gene. MSH3 is encoded by 222,341 base pairs and creates a protein consisting of 1137 amino acids. MSH3 is typically expressed at low levels in several transformed cell lines—including HeLa, K562, HL-60, and CEM—as well as a large range of normal tissues including spleen, thymus, prostate, testis, ovary, smal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hereditary Nonpolyposis Colorectal Cancer
Hereditary nonpolyposis colorectal cancer (HNPCC) is a hereditary predisposition to colon cancer. HNPCC includes (and was once synonymous with) Lynch syndrome, an autosomal dominant genetic condition that is associated with a high risk of colon cancer, endometrial cancer (second most common), ovary, stomach, small intestine, hepatobiliary tract, upper urinary tract, brain, and skin. The increased risk for these cancers is due to inherited genetic mutations that impair DNA mismatch repair. It is a type of cancer syndrome. Other HNPCC conditions include Lynch-like syndrome, polymerase proofreading-associated polyposis and familial colorectal cancer type X. Signs and symptoms Risk of cancer ''Lifetime risk and mean age at diagnosis for Lynch syndrome–associated cancers'' In addition to the types of cancer found in the chart above, it is understood that Lynch syndrome also contributes to an increased risk of small bowel cancer, pancreatic cancer, ureter/renal pelvis cancer, b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
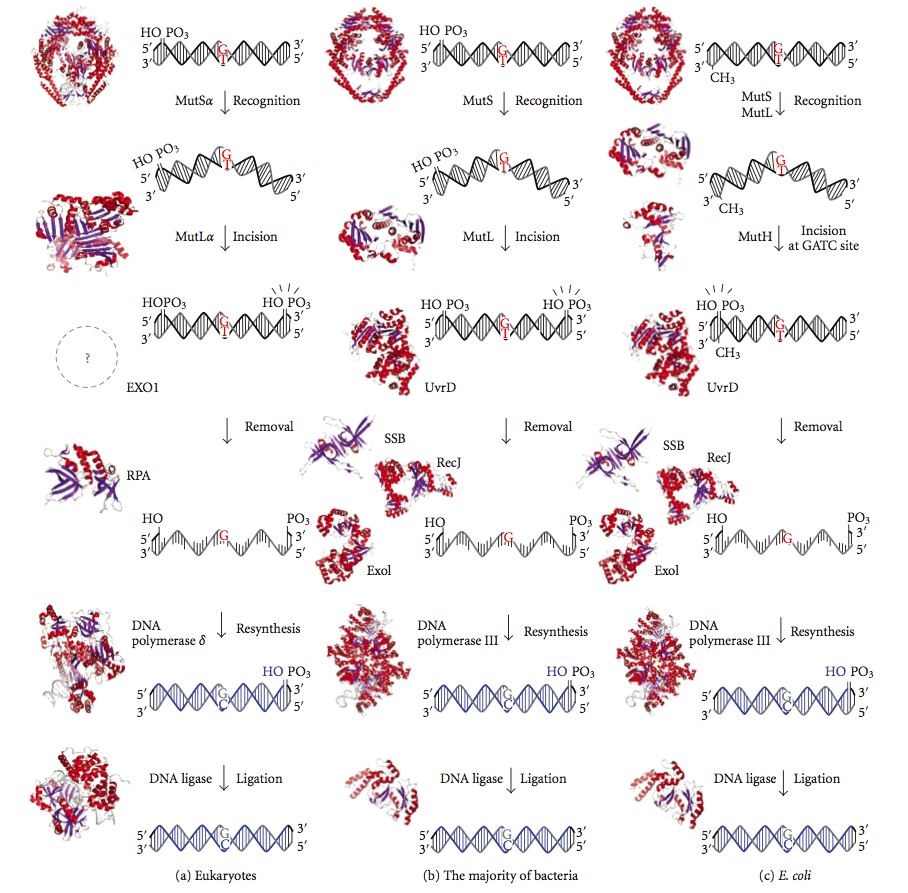
DNA Mismatch Repair
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of nucleobase, bases that can arise during DNA replication and Genetic recombination, recombination, as well as DNA repair, repairing some forms of DNA damage. Mismatch repair is strand-specific. During DNA synthesis the newly synthesised (daughter) strand will commonly include errors. In order to begin repair, the mismatch repair machinery distinguishes the newly synthesised strand from the template (parental). In gram-negative bacteria, transient methylase, hemimethylation distinguishes the strands (the parental is methylated and daughter is not). However, in other prokaryotes and eukaryotes, the exact mechanism is not clear. It is suspected that, in eukaryotes, newly synthesized lagging-strand DNA transiently contains Nick (DNA), nicks (before being sealed by DNA ligase) and provides a signal that directs mismatch proofreading systems to the appropriate st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
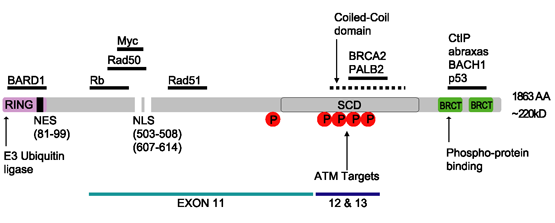
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a human tumor suppressor gene (also known as a caretaker gene) and is responsible for repairing DNA. ''BRCA1'' and ''BRCA2'' are unrelated proteins, but both are normally expressed in the cells of breast and other tissues, where they help repair damaged DNA, or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks. If ''BRCA1'' or ''BRCA2'' itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer. ''BRCA1'' and ''BRCA2'' have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a no ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tumor Suppressor Gene
A tumor suppressor gene (TSG), or anti-oncogene, is a gene that regulates a cell (biology), cell during cell division and replication. If the cell grows uncontrollably, it will result in cancer. When a tumor suppressor gene is mutated, it results in a loss or reduction in its function. In combination with other genetic mutations, this could allow the cell to grow abnormally. The Loss-of-function mutation, loss of function for these genes may be even more significant in the development of human cancers, compared to the activation of oncogenes. TSGs can be grouped into the following categories: caretaker genes, gatekeeper genes, and more recently landscaper genes. Caretaker genes ensure stability of the genome via DNA repair and subsequently when mutated allow mutations to accumulate. Meanwhile, gatekeeper genes directly regulate cell growth by either inhibiting cell cycle progression or inducing apoptosis. Lastly, landscaper genes regulate growth by contributing to the surrounding e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ataxia Telangiectasia And Rad3 Related
Serine/threonine-protein kinase ATR, also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1), is an enzyme that, in humans, is encoded by the ''ATR'' gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity. Function ATR is a serine/threonine-specific protein kinase that is involved in sensing DNA damage and activating the DNA damage checkpoint, leading to cell cycle arrest in eukaryotes. ATR is activated in response to persistent single-stranded DNA, which is a common intermediate formed during DNA damage detection and repair. Single-stranded DNA occurs at stalled replication forks and as an intermediate in DNA repair pathways such as nucleotide excision repair and homologous recombination repair. ATR is activated during more persistent issues with DNA damage ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 2
Chromosome 2 is one of the twenty-three pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 2 is the second-largest human chromosome, spanning more than 242 million base pairs and representing almost eight percent of the total DNA in human cells. Chromosome 2 contains the HOXD homeobox gene cluster. Fusion Humans have only twenty-three pairs of chromosomes, while all other extant members of Hominidae have twenty-four pairs. It is believed that Neanderthals and Denisovans had twenty-three pairs. Human chromosome 2 is a result of an end-to-end fusion of two ancestral chromosomes.It has been hypothesized that Human Chromosome 2 is a fusion of two ancestral chromosomes by Alec MacAndrew; accessed 18 May 2006. The ev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microsatellite Instability
Microsatellite instability (MSI) is the condition of genetic hypermutability (predisposition to mutation) that results from impaired DNA mismatch repair (MMR). The presence of MSI represents phenotypic evidence that MMR is not functioning normally. MMR corrects errors that spontaneously occur during DNA replication, such as single base mismatches or short insertions and deletions. The proteins involved in MMR correct polymerase errors by forming a complex that binds to the mismatched section of DNA, excises the error, and inserts the correct sequence in its place. Cells with abnormally functioning MMR are unable to correct errors that occur during DNA replication and consequently accumulate errors. This causes the creation of novel microsatellite fragments. Polymerase chain reaction-based assays can reveal these novel microsatellites and provide evidence for the presence of MSI. Microsatellites are repeated sequences of DNA. These sequences can be made of units of 1 to 6 bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |