|
Litchfieldia
''Litchfieldia'' is a genus of Gram-positive rod-shaped bacteria in the family Bacillaceae within the order Bacillales. The type species for this genus is ''Litchfieldia alkalitelluris.'' ''Litchfieldia'' is composed of species originally belonging to the genus ''Bacillus'', a genus that displayed extensive polyphyly due to the vague criteria used to assign new species to the genus. Studies using phylogenetic and comparative genomic analyses have clarified the taxonomy of ''Bacillus'' by restricting the genus to only include species closely related to ''Bacillus subtilis'' and ''Bacillus cereus'' as well as transfer many species into new novel genera such as '' Virgibacillus, Solibacillus, Brevibacillus'' and '' Ectobacillus''. The name ''Litchfieldia'' was named after the American marine microbiologist and ecologist Dr. Carol D. Litchfield (1936-2012). Biochemical characteristics and molecular signatures Source: Members of the genus ''Litchfieldia'' are aerobic or facultati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillaceae
Bacillaceae, from Latin "bacillus", meaning "little staff, wand", are a family of gram-positive, heterotrophic, rod-shaped bacteria that may produce endospores. Motile members of this family are characterized by peritrichous flagella. Some Bacillaceae are aerobic, while others are facultative or strict anaerobes. Most are not pathogenic, but ''Bacillus'' species are known to cause disease in humans. Gram-variable cell wall Some Bacillaceae, such as the genera '' Filobacillus, Lentibacillus,'' and '' Halobacillus'', stain Gram-negative or Gram-variable, but are known to have a Gram-positive cell wall.Lim, J.M., Jeon, C.O., Song, S.M., and C.J. Kim. 2005''Pontibacillus chungwhensis gen. nov., sp. nov., a moderately halophilic Gram-positive bacterium from a solar saltern in Korea'' Int. J. Syst. Evol. Microbiol. 55:165-170. Nomenclature Taxa within this family are sometimes colloquially identified as "bacilli". However, this term is ambiguous because it does not distinguish ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus
''Bacillus'', from Latin "bacillus", meaning "little staff, wand", is a genus of Gram-positive, rod-shaped bacteria, a member of the phylum ''Bacillota'', with 266 named species. The term is also used to describe the shape (rod) of other so-shaped bacteria; and the plural ''Bacilli'' is the name of the class of bacteria to which this genus belongs. ''Bacillus'' species can be either obligate aerobes which are dependent on oxygen, or facultative anaerobes which can survive in the absence of oxygen. Cultured ''Bacillus'' species test positive for the enzyme catalase if oxygen has been used or is present. ''Bacillus'' can reduce themselves to oval endospores and can remain in this dormant state for years. The endospore of one species from Morocco is reported to have survived being heated to 420 °C. Endospore formation is usually triggered by a lack of nutrients: the bacterium divides within its cell wall, and one side then engulfs the other. They are not true spores (i.e. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), classification and a taxonomic rank of an organism, as well as a unit of biodiversity. Other ways of defining species include their karyotype, DNA sequence, morphology (biology), morphology, behaviour, or ecological niche. In addition, palaeontologists use the concept of the chronospecies since fossil reproduction cannot be examined. The most recent rigorous estimate for the total number of species of eukaryotes is between 8 and 8.7 million. About 14% of these had been described by 2011. All species (except viruses) are given a binomial nomenclature, two-part name, a "binomen". The first part of a binomen is the name of a genus to which the species belongs. The second part is called the specific name (zoology), specific name or the specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidase
In biochemistry, an oxidase is an oxidoreductase (any enzyme that catalyzes a redox reaction) that uses dioxygen (O2) as the electron acceptor. In reactions involving donation of a hydrogen atom, oxygen is reduced to water (H2O) or hydrogen peroxide (H2O2). Some oxidation reactions, such as those involving monoamine oxidase or xanthine oxidase, typically do not involve free molecular oxygen. The oxidases are a subclass of the oxidoreductases. The use of dioxygen is the only unifying feature; in the EC classification, these enzymes are scattered in many categories. Examples An important example is EC 7.1.1.9 cytochrome c oxidase, the key enzyme that allows the body to employ oxygen in the generation of energy and the final component of the electron transfer chain. Other examples are: * EC 1.1.3.4 Glucose oxidase * EC 1.4.3.4 Monoamine oxidase * EC 1.14.-.- Cytochrome P450 oxidase * EC 1.6.3.1 NADPH oxidase * EC 1.17.3.2 Xanthine oxidase * EC 1.1.3.8 L-gulonolactone oxidase * EC 1. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an online database that maintains information on a proposed nomenclature of prokaryotes, following a phylogenomic approach based on a set of conserved single-copy proteins. In addition to resolving paraphyletic groups, this method also reassigns taxonomic ranks algorithmically, updating names in both cases. Information for archaea was added in 2020, along with a species classification based on average nucleotide identity. Each update incorporates new genomes as well as automated and manual curation of the taxonomy. An open-source tool called GTDB-Tk is available to classify draft genomes into the GTDB hierarchy. The GTDB system, via GTDB-Tk, has been used to catalogue not-yet-named bacteria in the human gut microbiome and other metagenomic sources. The GTDB is incorporated into the '' Bergey's Manual of Systematics of Archaea and Bacteria'' in 2019 as its phylogenomic resource. Methodology The genomes used to construct the phyloge ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In other words, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. In evolutionary biology, all life on Earth is theoretically part of a single phylogenetic tree, indicating common ancestry. Phylogenetics is the study of phylogenetic trees. The main challenge is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of species or taxa. Computational phylogenetics (also phylogeny inference) focuses on the algorithms involved in finding optimal phylogenetic tree in the phylogenetic landscape. Phylogenetic trees may be rooted or unrooted. In a ''rooted'' p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monophyly
In biological cladistics for the classification of organisms, monophyly is the condition of a taxonomic grouping being a clade – that is, a grouping of organisms which meets these criteria: # the grouping contains its own most recent common ancestor (or more precisely an ancestral population), i.e. excludes non-descendants of that common ancestor # the grouping contains all the descendants of that common ancestor, without exception Monophyly is contrasted with paraphyly and polyphyly as shown in the second diagram. A ''paraphyletic'' grouping meets 1. but not 2., thus consisting of the descendants of a common ancestor, excepting one or more monophyletic subgroups. A ''polyphyletic'' grouping meets neither criterion, and instead serves to characterize convergent relationships of biological features rather than genetic relationships – for example, night-active primates, fruit trees, or aquatic insects. As such, these characteristic features of a polyphyletic grouping are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
National Center For Biotechnology Information
The National Center for Biotechnology Information (NCBI) is part of the National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper. The NCBI houses a series of databases relevant to biotechnology and biomedicine and is an important resource for bioinformatics tools and services. Major databases include GenBank for DNA sequences and PubMed, a bibliographic database for biomedical literature. Other databases include the NCBI Epigenomics database. All these databases are available online through the Entrez search engine. NCBI was directed by David Lipman, one of the original authors of the BLAST sequence alignment program and a widely respected figure in bioinformatics. GenBank NCBI had responsibility for making available the GenBank DNA seque ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Prokaryotic Names With Standing In Nomenclature
List of Prokaryotic names with Standing in Nomenclature (LPSN) is an online database that maintains information on the naming and taxonomy of prokaryotes, following the taxonomy requirements and rulings of the International Code of Nomenclature of Prokaryotes. The database was curated from 1997 to June 2013 by Jean P. Euzéby. From July 2013 to January 2020, LPSN was curated by Aidan C. Parte. In February 2020, a new version of LPSN was published as a service of the Leibniz Institute DSMZ, thereby also integrating the Prokaryotic Nomenclature Up-to-date service and since 2022 LPSN is interconnected with the Type (Strain) Genome Server (TYGS), DSMZ's high-throughput platform for accurate genome-based taxonomy. See also * Code of Nomenclature of Prokaryotes Described from Sequence Data References External links List of Prokaryotic names with Standing in Nomenclature [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
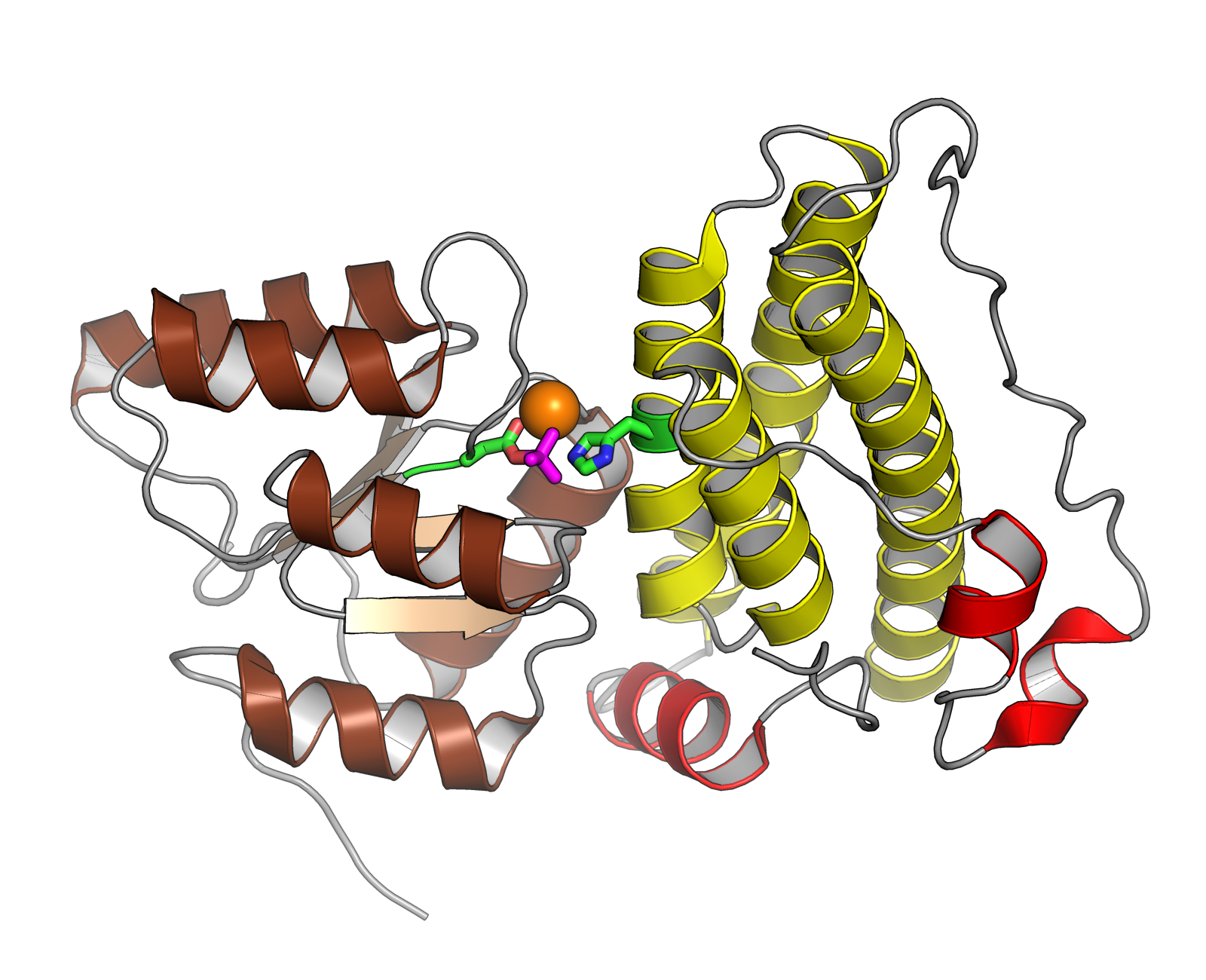
Response Regulator
In molecular biology, a response regulator is a protein that mediates a cell's response to changes in its environment as part of a two-component regulatory system. Response regulators are coupled to specific histidine kinases which serve as sensors of environmental changes. Response regulators and histidine kinases are two of the most common gene families in bacteria, where two-component signaling systems are very common; they also appear much more rarely in the genomes of some archaea, yeasts, filamentous fungi, and plants. Two-component systems are not found in metazoans. Function Response regulator proteins typically consist of a receiver domain and one or more effector domains, although in some cases they possess only a receiver domain and exert their effects through protein-protein interactions. In two-component signaling, a histidine kinase responds to environmental changes by autophosphorylation on a histidine residue, following which the response regulator receiver ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amidohydrolase
Amidohydrolases (or amidases) are a type of hydrolase that acts upon amide bonds. They are categorized under EC number EC 3.5.1 and 3.5.2. Examples include: * Beta-lactamase * Histone deacetylase * Urease The amidohydrolase superfamily is a large protein family of more than 20,000 members with diverse chemistry and physiologic roles. Due to its complexity and size, the amidohydrolase superfamily is being used by the Enzyme Function Initiative The Enzyme Function Initiative (EFI) is a large-scale collaborative project aiming to develop and disseminate a robust strategy to determine enzyme function through an integrated sequence–structure-based approach. The project was funded in May ... (EFI) for developing a large-scale strategy for functional assignment of unknown proteins. See also * EC 3.5.1 EC 3.5.2 Hydrolases {{3.5-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Metallopeptidase
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis. Most metalloproteases require zinc, but some use cobalt. The metal ion is coordinated to the protein via three ligands. The ligands coordinating the metal ion can vary with histidine, glutamate, aspartate, lysine, and arginine. The fourth coordination position is taken up by a labile water molecule. Treatment with chelating agents such as EDTA leads to complete inactivation. EDTA is a metal chelator that removes zinc, which is essential for activity. They are also inhibited by the chelator orthophenanthroline. Classification There are two subgroups of metalloproteinases: * Exopeptidases, metalloexopeptidases ( EC number: 3.4.17). * Endopeptidases, metalloendopeptidases (3.4.24). Well known metalloendopeptidases include ADAM prote ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |