|
Genome Skimming
Genome skimming is a sequencing approach that uses low-pass, shallow sequencing of a genome (up to 5%), to generate fragments of DNA, known as genome skims. These genome skims contain information about the high-copy fraction of the genome. The high-copy fraction of the genome consists of the ribosomal DNA, plastid genome (plastome), mitochondrial genome (mitogenome), and nuclear repeats such as microsatellites and transposable elements. It employs high-throughput, next generation sequencing technology to generate these skims. Although these skims are merely 'the tip of the genomic iceberg', phylogenomic analysis of them can still provide insights on evolutionary history and biodiversity at a lower cost and larger scale than traditional methods. Due to the small amount of DNA required for genome skimming, its methodology can be applied in other fields other than genomics. Tasks like this include determining the traceability of products in the food industry, enforcing international ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
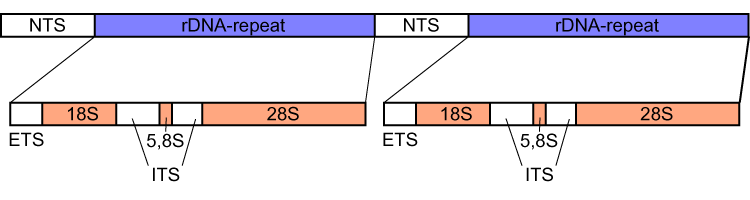
Internal Transcribed Spacer
Internal transcribed spacer (ITS) is the spacer DNA situated between the small-subunit ribosomal RNA (rRNA) and large-subunit rRNA genes in the chromosome or the corresponding transcribed region in the polycistronic rRNA precursor transcript. ITS across life domains In bacteria and archaea, there is a single ITS, located between the 16S and 23S rRNA genes. Conversely, there are two ITSs in eukaryotes: ITS1 is located between 18S and 5.8S rRNA genes, while ITS2 is between 5.8S and 28S (in opisthokonts, or 25S in plants) rRNA genes. ITS1 corresponds to the ITS in bacteria and archaea, while ITS2 originated as an insertion that interrupted the ancestral 23S rRNA gene. Organization In bacteria and archaea, the ITS occurs in one to several copies, as do the flanking 16S and 23S genes. When there are multiple copies, these do not occur adjacent to one another. Rather, they occur in discrete locations in the circular chromosome. It is not uncommon in bacteria to carry tRNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
16S RRNA (guanine1405-N7)-methyltransferase
16S rRNA (guanine1405-N7)-methyltransferase (, ''methyltransferase Sgm'', ''m7G1405 Mtase'', ''Sgm Mtase'', ''Sgm'', ''sisomicin-gentamicin methyltransferase'', ''sisomicin-gentamicin methylase'', ''GrmA'', ''RmtB'', ''RmtC'', ''ArmA'') is an enzyme with systematic name ''S-adenosyl-L-methionine:16S rRNA (guanine1405-N7)-methyltransferase''. This enzyme catalyses the following chemical reaction : S-adenosyl-L-methionine + guanine1405 in 16S rRNA 16S rRNA may refer to: * 16S ribosomal RNA 16 S ribosomal RNA (or 16 S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome ( SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure. The g ... \rightleftharpoons S-adenosyl-L-homocysteine + 7-methylguanine1405 in 16S rRNA The enzyme specifically methylates guanine1405 at N7 in 16S rRNA. References External links * {{Portal bar, Biology, border=no EC 2.1.1 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NADH2 Dehydrogenase (ubiquinone)
NADH2 dehydrogenase (ubiquinone) may refer to: * NADH dehydrogenase NADH dehydrogenase is an enzyme that converts nicotinamide adenine dinucleotide (NAD) from its reduced form (NADH) to its oxidized form (NAD+). Members of the NADH dehydrogenase family and analogues are commonly systematically named using the for ... * NADH:ubiquinone reductase (non-electrogenic) {{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. Horizontal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-Mendelian Inheritance
Non-Mendelian inheritance is any pattern in which traits do not segregate in accordance with Mendel's laws. These laws describe the inheritance of traits linked to single genes on chromosomes in the nucleus. In Mendelian inheritance, each parent contributes one of two possible alleles for a trait. If the genotypes of both parents in a genetic cross are known, Mendel's laws can be used to determine the distribution of phenotypes expected for the population of offspring. There are several situations in which the proportions of phenotypes observed in the progeny do not match the predicted values. Non-Mendelian inheritance plays a role in several disease which affected the processes. Types Incomplete dominance In cases of intermediate inheritance due to incomplete dominance, the principle of dominance discovered by Mendel does not apply. Nevertheless, the principle of uniformity works, as all offspring in the F1-generation have the same genotype and same phenotype. Mendel's pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Marker
A molecular marker is a molecule, sampled from some source, that gives information about its source. For example, DNA is a molecular marker that gives information about the organism from which it was taken. For another example, some proteins can be molecular markers of Alzheimer's disease in a person from which they are taken. Molecular markers may be non-biological. Non-biological markers are often used in environmental studies. Genetic markers In genetics, a molecular marker (identified as genetic marker) is a fragment of DNA that is associated with a certain location within the genome. Molecular markers are used in molecular biology and biotechnology to identify a particular sequence of DNA in a pool of unknown DNA. Types of genetic markers There are many types of genetic markers, each with particular limitations and strengths. Within genetic markers there are three different categories: "First Generation Markers", "Second Generation Markers", and "New Generation Marker ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-nucleotide Polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with incr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Barcoding
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called " sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries. Different gene regions are used to identify the different organismal groups using barcoding. The most commonly used barcode region for animals and some protists is a portion of the cytochrome ''c'' oxidase I (COI or COX1) gene, found in mitochondrial DNA. Other genes suitable for DNA barcodin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sanger Sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Frederick Sanger and colleagues in 1977, it became the most widely used sequencing method for approximately 40 years. It was first commercialized by Applied Biosystems in 1986. More recently, higher volume Sanger sequencing has been replaced by next generation sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use for smaller-scale projects and for validation of deep sequencing results. It still has the advantage over short-read sequencing technologies (like Illumina) in that it can produce DNA sequence reads of > 500 nucleotides and maintains a very low error rate with accuracies around 99.99%. Sanger sequencing is still actively being used in efforts for public health initiati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plastid
The plastid (Greek: πλαστός; plastós: formed, molded – plural plastids) is a membrane-bound organelle found in the cells of plants, algae, and some other eukaryotic organisms. They are considered to be intracellular endosymbiotic cyanobacteria. Examples include chloroplasts (used for photosynthesis), chromoplasts (used for pigment synthesis and storage), and leucoplasts (non-pigmented plastids that can sometimes differentiate). The event which led to permanent endosymbiosis in the Archaeplastida clade (of land plants, red algae, and green algae) probably occurred with a cyanobiont (a symbiotic cyanobacteria) related to the genus '' Gloeomargarita'', around 1.5 billion years ago. A later primary endosymbiosis event occurred in photosynthetic ''Paulinella'' amoeboids about 90–140 million years ago. This plastid belongs to the "PS-clade" (of the cyanobacteria genera ''Prochlorococcus'' and ''Synechococcus''). Secondary and tertiary endosymbiosis has also occurred, in a wid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coverage (genetics)
Coverage (or depth) in DNA sequencing is the number of unique reads that include a given nucleotide in the reconstructed sequence. Deep sequencing refers to the general concept of aiming for high number of unique reads of each region of a sequence. Rationale Even though the sequencing accuracy for each individual nucleotide is very high, the very large number of nucleotides in the genome means that if an individual genome is only sequenced once, there will be a significant number of sequencing errors. Furthermore, many positions in a genome contain rare single-nucleotide polymorphisms (SNPs). Hence to distinguish between sequencing errors and true SNPs, it is necessary to increase the sequencing accuracy even further by sequencing individual genomes a large number of times. Ultra-deep sequencing The term "ultra-deep" can sometimes also refer to higher coverage (>100-fold), which allows for detection of sequence variants in mixed populations. In the extreme, error-corrected sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |