|
FAM71F2
FAM71F2 or Family with Sequence Similarity 71 member F2 is a protein that in humans is encoded by the ''Family with Sequence Similarity 71 member F2 gene''. This gene is highly active in the reproductive tissues, specifically the testis, and may serve as a potential biomarker for determining metastatic testicular cancer. Gene Location FAM71F2 gene is located on chromosome 7 in humans (7q32.1), starting at 128,671,636 and ending at 128,702,262 on the Sense (molecular biology), positive strand. The gene paralog FAM71F1 and the gene LINC01000 directly neighbor FAM71F2 on chromosome 7. Size of gene The gene spans 30,627 base pairs and codes for 12 exons. Common aliases FAM71F2 is also referred to as family with sequence similarity 137 member B, FAM137B. mRNA FAM71F2 has 14 transcript variants. ''Isoform'' ''a'' is the longest of the mRNA transcripts and spans 5,775 base pairs that translates into a 309 amino acids sequence. It codes for 5 exons. Other alternative splic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 7
Chromosome 7 is one of the 23 pairs of chromosomes in humans, who normally have two copies of this chromosome. Chromosome 7 spans about 160 million base pairs (the building material of DNA) and represents between 5 and 5.5 percent of the total DNA in cells. Genes Number of genes The following are some of the gene count estimates of human chromosome 7. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project ( CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes. Gene list The following is a partial list of genes on human chromosome 7. For complete list, see the link in the infobox on the right. Diseases and disorders The following diseases are some of those related to gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metabolic reactions, DNA replication, Cell signaling, responding to stimuli, providing Cytoskeleton, structure to cells and Fibrous protein, organisms, and Intracellular transport, transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the Nucleic acid sequence, nucleotide sequence of their genes, and which usually results in protein folding into a specific Protein structure, 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called pep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reptile
Reptiles, as commonly defined, are a group of tetrapods with an ectothermic metabolism and Amniotic egg, amniotic development. Living traditional reptiles comprise four Order (biology), orders: Testudines, Crocodilia, Squamata, and Rhynchocephalia. About 12,000 living species of reptiles are listed in the Reptile Database. The study of the traditional reptile orders, customarily in combination with the study of modern amphibians, is called herpetology. Reptiles have been subject to several conflicting Taxonomy, taxonomic definitions. In Linnaean taxonomy, reptiles are gathered together under the Class (biology), class Reptilia ( ), which corresponds to common usage. Modern Cladistics, cladistic taxonomy regards that group as Paraphyly, paraphyletic, since Genetics, genetic and Paleontology, paleontological evidence has determined that birds (class Aves), as members of Dinosauria, are more closely related to living crocodilians than to other reptiles, and are thus nested among re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetylation Site
: In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite reaction, the removal of an acetyl group from a chemical compound. Acetylation/deacetylation in biology Histone deacetylases "play crucial roles in gene transcription and most likely in all eukaryotic biological processes that involve chromatin". Acetylation is one type of post-translational modification of proteins. The acetylation of the ε-amino group of lysine, which is common, converts a charged side chain to a neutral one. Acetylation/deacetylation of histones also plays a role in gene expression and cancer. These modifications are effected by enzymes called histone acetyltransferases (HATs) and histone deacetylases (HDACs). Two general mechanisms are known for deacetylation. One mechanism involves zinc binding to the acetyl oxyg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphorylated Site
Protein phosphorylation is a reversible post-translational modification of proteins in which an amino acid residue is phosphorylated by a protein kinase by the addition of a covalently bound phosphate group. Phosphorylation alters the structural conformation of a protein, causing it to become activated, deactivated, or otherwise modifying its function. Approximately 13,000 human proteins have sites that are phosphorylated. The reverse reaction of phosphorylation is called dephosphorylation, and is catalyzed by protein phosphatases. Protein kinases and phosphatases work independently and in a balance to regulate the function of proteins. The amino acids most commonly phosphorylated are serine, threonine, tyrosine, and histidine. These phosphorylations play important and well-characterized roles in signaling pathways and metabolism. However, other amino acids can also be phosphorylated post-translationally, including arginine, lysine, aspartic acid, glutamic acid and cysteine, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helices
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid that is four residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery In the early 1930s, William Astbury showed that there were dras ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
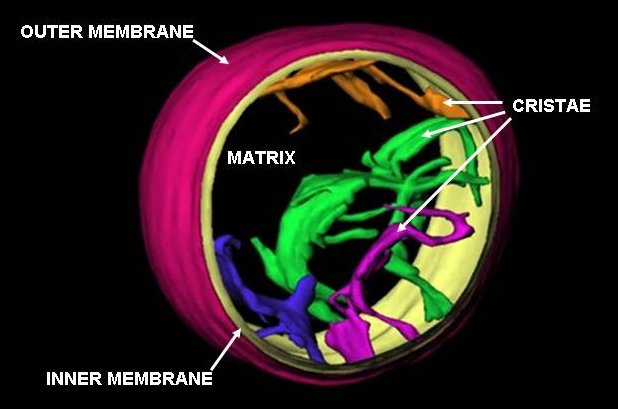
Mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'', meaning a thread-like granule, was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 ''Scientific American'' article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal '' Henneguya salminicola'' is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome. A large number of unicellular organisms, such as microspo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
R-2 Mitochondrial Pre-sequence Cleavage Site
R- may represent: *a type of chirality, in chemical notation *an ''R'' prefix used for various constants *the set of negative real numbers *negative reinforcement In behavioral psychology, reinforcement refers to consequences that increase the likelihood of an organism's future behavior, typically in the presence of a particular '' antecedent stimulus''. For example, a rat can be trained to push a lever ..., in behavioural psychology *membership of the United States Republican Party, when placed before a state abbreviation, usually in parentheses after the name of a politician {{Disambiguation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes. The eukaryotes emerged within the archaeal kingdom Promethearchaeati and its sole phylum Promethearchaeota. This implies that there are only two domains of life, Bacteria and Archaea, with eukaryotes incorporated among the Archaea. Eukaryotes first emerged during the Paleoproterozoic, likely as flagellated cells. The leading evolutionary theory is they were created by symbiogenesis between an anaerobic Promethearchaeati archaean and an aerobic proteobacterium, which form ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |