|
Endoribonuclease
In biochemistry, an endoribonuclease is a class of enzyme which is a type of ribonuclease (an RNA cleaver), itself a type of endonuclease (a nucleotide cleaver). It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Example includes both single proteins such as RNase III, RNase A, RNase T1, RNase T2 and RNase H and also complexes of proteins with RNA such as RNase P and the RNA-induced silencing complex. Further examples include endoribonuclease XendoU found in frogs (''Xenopus ''Xenopus'' () (Gk., ξενος, ''xenos'' = strange, πους, ''pous'' = foot, commonly known as the clawed frog) is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described with ...''). External links * {{Nucleases EC 3.1 Ribonucleases ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endoribonuclease XendoU
In molecular biology, Endoribonuclease XendoU refers to a protein domain. This particular entry represents endoribonucleases involved in RNA biosynthesis which has been named XendoU in ''Xenopus laevis'' (African clawed frog). This protein domain belongs to a family of evolutionarily related proteins. XendoU is a U-specific metal dependent enzyme that produces products with a 2' -3' cyclic phosphate termini. Function The endonuclease, XendoU, is highly involved in the biosynthesis of a specific subclass of ''Xenopus laevis'' encoded small nucleolar RNAs (snoRNA) which are a large family of non-coding RNAs with essential roles in ribosome biogenesis. Most snoRNAs are encoded in introns and are released through the splicing reaction. Others, instead, produced by an alternative pathway consisting of endonucleolytic processing of pre-mRNA. XendoU, is the endoribonuclease responsible for this activity. The XendoU-RNA complex is manganese (Mn2+)-independent. This infers that RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biochemistry
Biochemistry, or biological chemistry, is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology, and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all List of life sciences, areas of the life sciences are being uncovered and developed through biochemical methodology and research.#Voet, Voet (2005), p. 3. Biochemistry focuses on understanding the chemical basis that allows biomolecule, biological molecules to give rise to the processes that occur within living Cell (biology), cells and between cells,#Karp, Karp (2009), p. 2. in turn relating greatly to the understanding of tissue (biology), tissues and organ (anatomy), organs as well as organism structure and function.#Miller, Miller (2012). p. 62. Biochemistry is closely ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while many, typically called '' restriction endonucleases'' or ''restriction enzymes'', cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (''endo'') portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the ''restriction site''. Typically, a restriction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all Life, life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common Nutrient, nutrients by the liver. Nucleotides are composed of three subunit molecules: a nucleobase, a pentose, five-carbon sugar (ribose or deoxyribose), and a phosphate group consisting of one to three phosphates. The four nucleobases in DNA are guanine, adenine, cytosine, and thymine; in RNA, uracil is used in place of thymine. Nucleotides also play a central role in metabolism at a fundamental, cellular level. They provide chemical energy—in the form of the nucleoside triphosphates, adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP), and uridine triph ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase III
Ribonuclease III (RNase III or RNase C) (BREND3.1.26.3 is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the ''pnp'' autoregulatory mechanism. Types of RNase III The RNase III superfamily is divided into four known classes: 1, 2, 3, and 4. Each class is defined by its domain structure.Liang Y-H, Lavoie M, Comeau M-A, Elela SA, Ji X. Structure of a Eukaryotic RNase III Post-Cleavage Complex Reveals a Double- Ruler Mechanism for Substrate Selection. Molecular cell. 2014;54(3):431-444. doi:10.1016/j.molcel.2014.03.006. Class 1 RNase III *Class 1 RNase III enzymes have a homodimeric structure whose function is to cleave dsRNA into multipl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase A
Pancreatic ribonuclease family (, ''RNase'', ''RNase I'', ''RNase A'', ''pancreatic RNase'', ''ribonuclease I'', ''endoribonuclease I'', ''ribonucleic phosphatase'', ''alkaline ribonuclease'', ''ribonuclease'', ''gene S glycoproteins'', ''Ceratitis capitata alkaline ribonuclease'', ''SLSG glycoproteins'', ''gene S locus-specific glycoproteins'', ''S-genotype-assocd. glycoproteins'', ''ribonucleate 3'-pyrimidino-oligonucleotidohydrolase'') is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles. Specifically, the enzymes are involved in endonucleolytic cleavage of 3'-phosphomononucleotides and 3'-phosphooligonucleotides ending in C-P or U-P with 2',3'-cyclic phosphate intermediates. Ribonuclease can unwind the RNA helix by complexing with single-stranded RNA; the complex arises by an extended multi-site cation-anion interaction between lysine and arginine residues of the enzyme and phosphate groups of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase T1
Ribonuclease T1 (, ''guanyloribonuclease'', ''Aspergillus oryzae ribonuclease'', ''RNase N1'', ''RNase N2'', ''ribonuclease N3'', ''ribonuclease U1'', ''ribonuclease F1'', ''ribonuclease Ch'', ''ribonuclease PP1'', ''ribonuclease SA'', ''RNase F1'', ''ribonuclease C2'', ''binase'', ''RNase Sa'', ''guanyl-specific RNase'', ''RNase G'', ''RNase T1'', ''ribonuclease guaninenucleotido-2'-transferase (cyclizing)'', ''ribonuclease N3'', ''ribonuclease N1'') is a fungal endonuclease that cleaves single-stranded RNA after guanine residues, i.e., on their 3' end; the most commonly studied form of this enzyme is the version found in the mold ''Aspergillus oryzae''. Owing to its specificity for guanine, RNase T1 is often used to digest denatured RNA prior to sequencing. Similar to other ribonucleases such as barnase and RNase A, ribonuclease T1 has been popular for folding studies. Structurally, ribonuclease T1 is a small α+β protein (104 amino acids) with a four-stranded, antiparall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase T2
Ribonuclease T2 is an enzyme that in humans is encoded by the ''RNASET2'' gene. It is a type of endoribonuclease In biochemistry, an endoribonuclease is a class of enzyme which is a type of ribonuclease (an RNA cleaver), itself a type of endonuclease (a nucleotide cleaver). It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Ex .... This ribonuclease gene is a novel member of the Rh/T2/S-glycoprotein class of extracellular ribonucleases. It is a single copy gene that maps to 6q27, a region associated with human malignancies and chromosomal rearrangement. RNASET2 has been reported as a tumour associated antigen in anaplastic large cell lymphoma and other lymphomas. References Further reading * * * * * * * * * {{gene-6-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
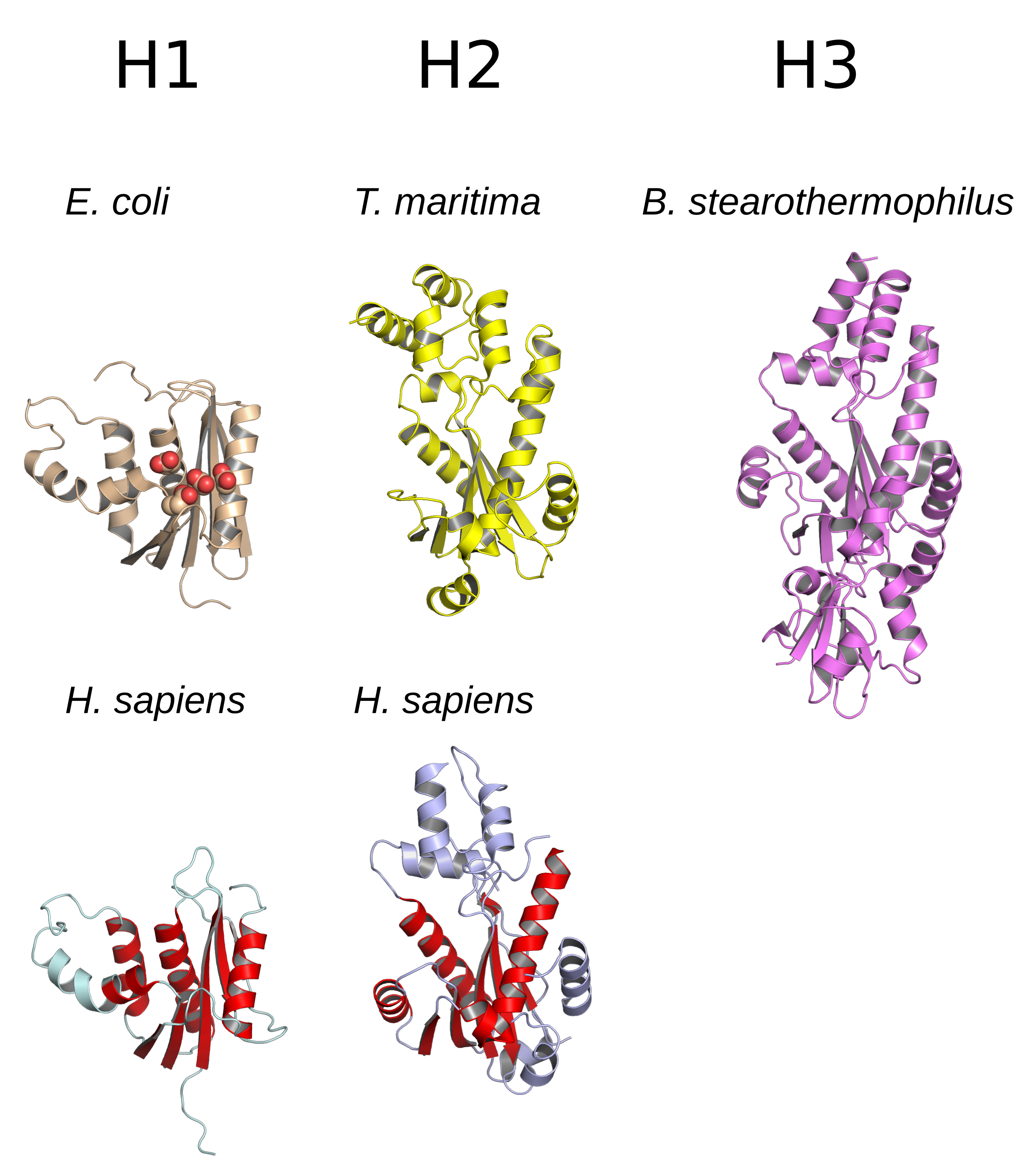
RNase H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-nucleotide sequence, sequence-specific endonuclease enzymes that catalysis, catalyze the cleavage of RNA in an RNA/DNA substrate (chemistry), substrate via a hydrolysis, hydrolytic chemical mechanism, mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate (chemistry), substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reve ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNase P
Ribonuclease P (, ''RNase P'') is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature (the other being the ribosome), the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Its best characterised enzyme activity is the generation of mature 5′-ends of tRNAs by cleaving the 5′-leader elements of precursor-tRNAs. Cellular RNase Ps are ribonucleoproteins. The RNA from bacterial RNase P retains its catalytic activity in the ab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |