|
Eggerthellaceae Phylogenetic Relationships
The ''Eggerthellaceae'' are a family of Gram-positive, rod- or coccus-shaped Actinomycetota. It is the sole family within the order ''Eggerthellales.'' The name ''Eggerthellaceae'' is derived from the Latin term ''Eggerthella,'' referring to the type genus of the family and the suffix "-ceae," an ending used to denote a family. Together, ''Eggerthellaceae'' refers to a family whose nomenclatural type is the genus ''Eggerthella''. Biochemical characteristics and molecular signatures Members of this family are mostly anaerobic, non-motile (with the exception of some ''Gordonibacter'' and ''Senegalimassilia'' species that exhibit motility), asaccharolytic and do not form spores. ''Eggerthellaceae'' species are commonly isolated from human and animal faeces and other human sources such as the colon, vagina, oral cavity and blood. Some species have also been isolated from human samples of periodontal or endodontic infections, Crohn's disease and severe blood bacteremia. The G+C co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Outgroup (cladistics)
In cladistics or phylogenetics, an outgroup is a more distantly related group of organisms that serves as a reference group when determining the evolutionary relationships of the ingroup, the set of organisms under study, and is distinct from sociological outgroups. The outgroup is used as a point of comparison for the ingroup and specifically allows for the phylogeny to be rooted. Because the polarity (direction) of character change can be determined only on a rooted phylogeny, the choice of outgroup is essential for understanding the evolution of traits along a phylogeny. History Although the concept of outgroups has been in use from the earliest days of cladistics, the term "outgroup" is thought to have been coined in the early 1970s at the American Museum of Natural History. Prior to the advent of the term, various other terms were used by evolutionary biologists, including "exgroup", "related group", and "outside groups". Choice of outgroup The chosen outgroup is hypothe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Eggerthellaceae Phylogenetic Relationships
The ''Eggerthellaceae'' are a family of Gram-positive, rod- or coccus-shaped Actinomycetota. It is the sole family within the order ''Eggerthellales.'' The name ''Eggerthellaceae'' is derived from the Latin term ''Eggerthella,'' referring to the type genus of the family and the suffix "-ceae," an ending used to denote a family. Together, ''Eggerthellaceae'' refers to a family whose nomenclatural type is the genus ''Eggerthella''. Biochemical characteristics and molecular signatures Members of this family are mostly anaerobic, non-motile (with the exception of some ''Gordonibacter'' and ''Senegalimassilia'' species that exhibit motility), asaccharolytic and do not form spores. ''Eggerthellaceae'' species are commonly isolated from human and animal faeces and other human sources such as the colon, vagina, oral cavity and blood. Some species have also been isolated from human samples of periodontal or endodontic infections, Crohn's disease and severe blood bacteremia. The G+C co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-directed RNA Polymerase
A polymerase is an enzyme (Enzyme Commission number, EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using Base pair, base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophile, thermophilic bacteria, bacterium, ''Thermus aquaticus'' (''Taq'') (Protein Data Bank, PDB]1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template dependent or template independent. Polynucleotide adenylyltransferase, Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. Types By function *DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol POLG, γ, POLQ, θ, DNA polymera ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UDP-glucose 4-epimerase
The enzyme UDP-glucose 4-epimerase (), also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity. Additionally, human and some bacterial GALE isoforms reversibly catalyze the formation of UDP-''N''-acetylgalactosamine (UDP-GalNAc) from UDP-''N''-acetylglucosamine (UDP-GlcNAc) in the presence of NAD+, an initial step in glycoprotein or glycolipid synthesis. Historical significance Dr. Luis Leloir deduced the role of GALE in galactose metabolism during his tenure at the Instituto de Investigaciones Bioquímicas del Fundación Campomar, initially terming the enzyme waldenase. Dr. Leloir was awarded the 1970 Nobel Prize in Chemistry for his discover ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
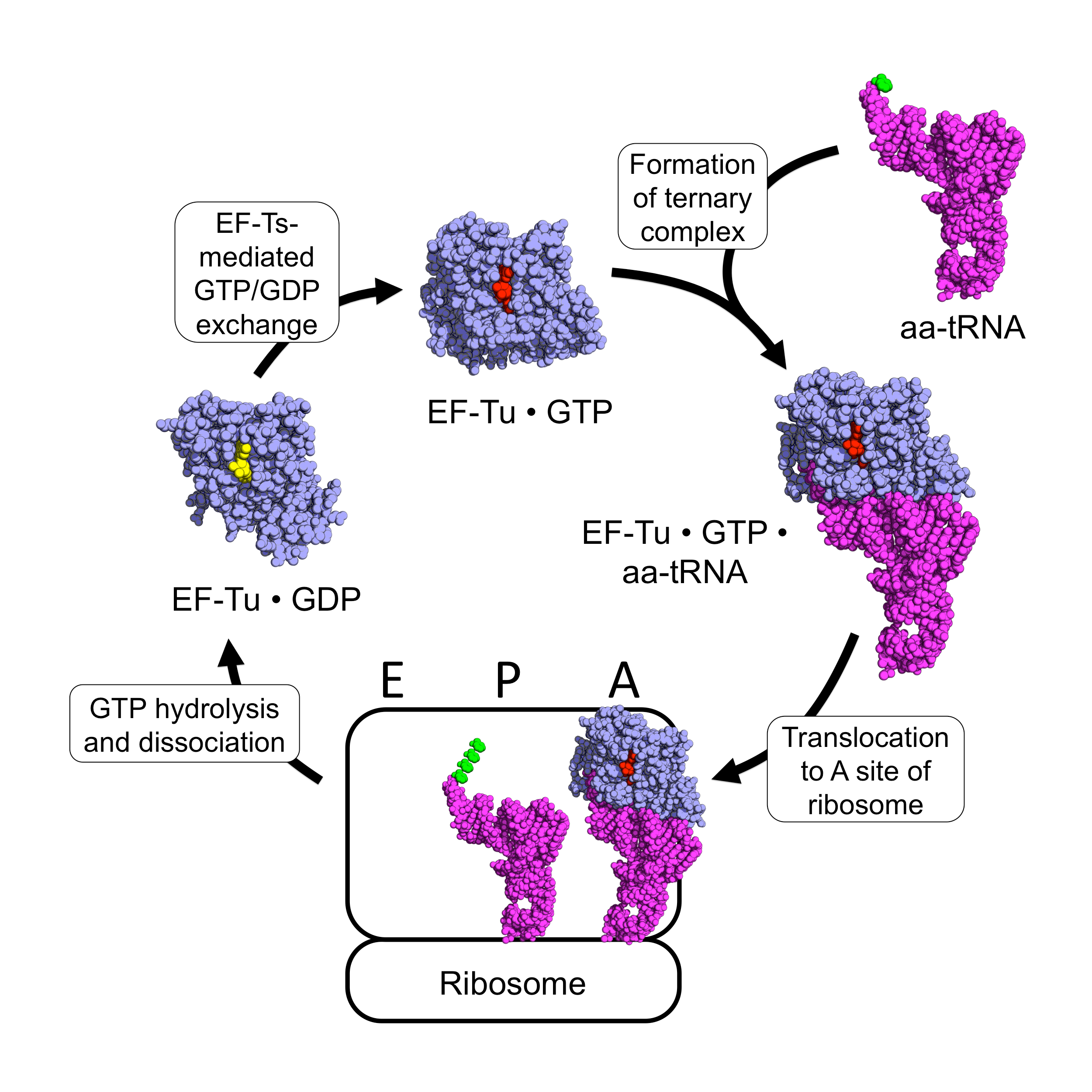
Elongation Factor Tu
EF-Tu (elongation factor thermo unstable) is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochondria as TUFM. As a family of elongation factors, EF-Tu also includes its eukaryotic and archaeal homolog, the alpha subunit of eEF-1 (EF-1A). Background Elongation factors are part of the mechanism that synthesizes new proteins through translation in the ribosome. Transfer RNAs (tRNAs) carry the individual amino acids that become integrated into a protein sequence, and have an anticodon for the specific amino acid that they are charged with. Messenger RNA (mRNA) carries the genetic information that encodes the primary structure of a pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease II
Ribonuclease II may refer to one of two enzymes: *Ribonuclease T2 Ribonuclease T2 (, ''ribonuclease II'', ''base-non-specific ribonuclease'', ''nonbase-specific RNase'', ''RNase (non-base specific)'', ''non-base specific ribonuclease'', ''nonspecific RNase'', ''RNase Ms'', ''RNase M'', ''RNase II'', ''Escherich ... * Exoribonuclease II {{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase III Holoenzyme
DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg (son of Arthur Kornberg) and Malcolm Gefter in 1970. The complex has high processivity (i.e. the number of nucleotides added per binding event) and, specifically referring to the replication of the ''E.coli'' genome, works in conjunction with four other DNA polymerases (Pol I, Pol II, Pol IV, and Pol V). Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity reading 3'→5' and synthesizing 5'→3'. DNA Pol III is a component of the replisome, which is located at the replication fork. Components The replisome is composed of the following: *2 DNA Pol III enzymes, each comprising α, ε and θ subunits. (It has been proven that there is a third copy of Pol III at the replisome.) **the α subunit (encoded b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conserved Signature Indels
Conserved signature inserts and deletions (CSIs) in protein sequences provide an important category of molecular markers for understanding phylogenetic relationships. CSIs, brought about by rare genetic changes, provide useful phylogenetic markers that are generally of defined size and they are flanked on both sides by conserved regions to ensure their reliability. While indels can be arbitrary inserts or deletions, CSIs are defined as only those protein indels that are present within conserved regions of the protein. The CSIs that are restricted to a particular clade or group of species, generally provide good phylogenetic markers of common evolutionary descent. Due to the rarity and highly specific nature of such changes, it is less likely that they could arise independently by either convergent or parallel evolution (i.e. homoplasy) and therefore are likely to represent synapomorphy. Other confounding factors such as differences in evolutionary rates at different sites or amon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gram-positive Bacteria
In bacteriology, gram-positive bacteria are bacteria that give a positive result in the Gram stain test, which is traditionally used to quickly classify bacteria into two broad categories according to their type of cell wall. Gram-positive bacteria take up the crystal violet stain used in the test, and then appear to be purple-coloured when seen through an optical microscope. This is because the thick peptidoglycan layer in the bacterial cell wall retains the stain after it is washed away from the rest of the sample, in the decolorization stage of the test. Conversely, gram-negative bacteria cannot retain the violet stain after the decolorization step; alcohol used in this stage degrades the outer membrane of gram-negative cells, making the cell wall more porous and incapable of retaining the crystal violet stain. Their peptidoglycan layer is much thinner and sandwiched between an inner cell membrane and a bacterial outer membrane, causing them to take up the counterstain ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Slackia
''Slackia'' is a genus of Actinomycetota, in the family Coriobacteriaceae. ''Slackia'' is named after the microbiologist Geoffrey Slack Geoffrey, Geoffroy, Geoff, etc., may refer to: People * Geoffrey (name), including a list of people with the name * Geoffroy (surname), including a list of people with the name * Geoffrey of Monmouth (c. 1095–c. 1155), clergyman and one of the .... References External links LPSN Coriobacteriaceae Bacterial vaginosis Bacteria genera {{actinobacteria-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




.jpg)