|
Deoxyribonucleases
Deoxyribonuclease (DNase, for short) refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells includes breaking down extracellular DNA (ecDNA) excreted by apoptosis, necrosis, and neutrophil extracellular traps (NET) of cells to help reduce inflammatory responses that otherwise are elicited. A wide variety of deoxyribonucleases are known and fall into one of two families ( DNase I or DNase II), which differ in their substrate specificities, chemical mechanisms, and biological functions. Laboratory applications of DNase include purifying proteins when extracted from prokaryotic organisms. Additionally, DNase has been applied as a treatment for diseases that are caused by ecDNA in the blood plasma. Assays of DNase are emerging in the research field as well. Types The two main types of DNase found in animals are known as deoxyr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exodeoxyribonuclease
Exodeoxyribonucleases are both exonucleases and deoxyribonucleases. They catalyze digestion of the ends of linear DNA. They are a type of esterase. They are classified EC 3.1.11. See also * Deoxyribonuclease Deoxyribonuclease (DNase, for short) refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells ... External links * EC 3.1 Deoxyribonucleases {{3.1-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNase MOA
Deoxyribonuclease (DNase, for short) refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells includes breaking down extracellular DNA (ecDNA) excreted by apoptosis, necrosis, and neutrophil extracellular traps (NET) of cells to help reduce inflammatory responses that otherwise are elicited. A wide variety of deoxyribonucleases are known and fall into one of two families ( DNase I or DNase II), which differ in their substrate specificities, chemical mechanisms, and biological functions. Laboratory applications of DNase include purifying proteins when extracted from prokaryotic organisms. Additionally, DNase has been applied as a treatment for diseases that are caused by ecDNA in the blood plasma. Assays of DNase are emerging in the research field as well. Types The two main types of DNase found in animals are known as deoxyr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
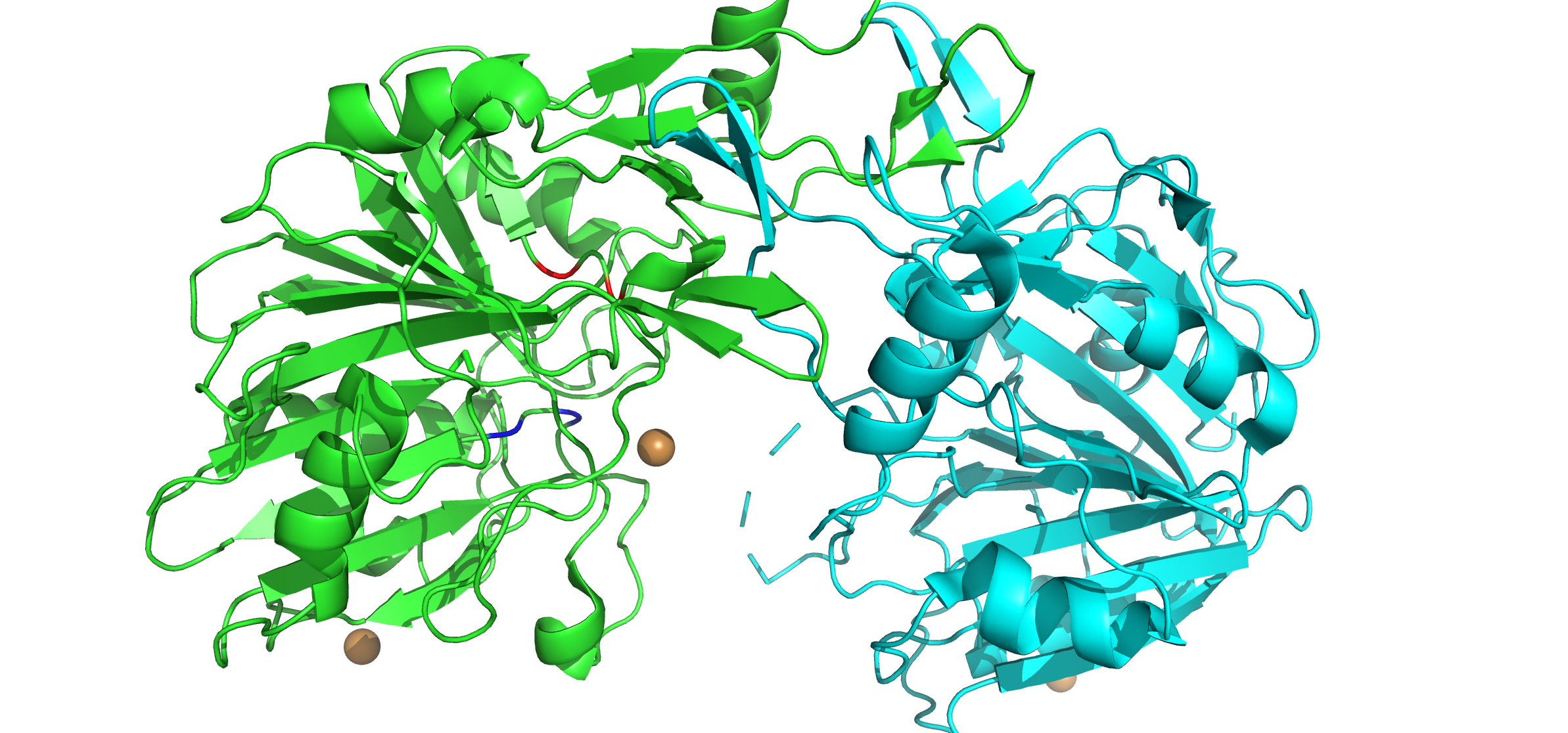
DNase 1 Image 1
Deoxyribonuclease (DNase, for short) refers to a group of glycoprotein endonucleases which are enzymes that catalyze the Hydrolysis, hydrolytic cleavage of Phosphodiester bonds, phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells includes breaking down extracellular DNA (ecDNA) excreted by apoptosis, necrosis, and neutrophil extracellular traps (NET) of cells to help reduce inflammatory responses that otherwise are elicited. A wide variety of deoxyribonucleases are known and fall into one of two families (Deoxyribonuclease I, DNase I or Deoxyribonuclease II, DNase II), which differ in their Substrate (biochemistry), substrate specificities, chemical mechanisms, and biological functions. Laboratory applications of DNase include Protein purification, purifying proteins when extracted from Prokaryote, prokaryotic organisms. Additionally, DNase has been applied as a treatment for diseases that are caused by ecDNA in the blood plasma. Ass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endodeoxyribonuclease
In biochemistry, an endodeoxyribonuclease is a class of enzyme which is a type of deoxyribonuclease (a DNA cleaver), itself a type of endonuclease (a nucleotide cleaver). They catalyze cleavage of the phosphodiester bonds in DNA. They are classified with EC numbers 3.1.21 through 3.1.25. Examples include: * DNA restriction enzymes * micrococcal nuclease See also * Ribonuclease Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within th ... * UvrABC endonuclease External links * EC 3.1 Deoxyribonucleases {{hydrolase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease. In both archaea and eukaryotes, one of the main routes of RNA degradation is performed by the multi-protein exosome complex, which consists largely of 3′ to 5′ exoribonucleases. Significance to polymerase RNA polymerase II is known to be in effect during transcriptional termination; it works with a 5' exonuclease (human gene Xrn2) to degrade the newly formed transcr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNASE1L3
Deoxyribonuclease gamma (also termed DNase γ, deoxyribonuclease 1L3, DNASE1L3, of deoxyribonuclease I like 3) is an enzyme that in humans is encoded by the ''DNASE1L3'' (also termed the ''deoxyribonuclease 1L3'' or ''deoxyribonuclease 1 like 3'') gene. This gene's is located on chromosome 3's " p arm", i.e., short arm, between region 1, band 4, sub-band 3 and region 2, band 1, sub-band 1 (this location's is abbreviation as 3p14.3-p21.1) Function DNASE1L3 belongs to the family of deoxyribonuclease enzymes that are responsible for degrading DNA. Specifically, DNASE1L3 plays a key role in the breakdown of extracellular DNA, particularly DNA released from dying cells due to apoptosis or necrosis. This function is important for maintaining cellular homeostasis and preventing the accumulation of DNA debris, which could otherwise trigger widespread inflammatory autoimmune responses. Clinical significance Role in autoimmune response Humans with inactivating mutations in both ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNase II
Deoxyribonuclease II (, ''DNase II'', ''pancreatic DNase II'', ''deoxyribonucleate 3'-nucleotidohydrolase'', ''pancreatic DNase II'', ''acid deoxyribonuclease'', ''acid DNase'') is an endonuclease that hydrolyzes phosphodiester linkages of deoxyribonucleotide in native and denatured DNA, yielding products with 3'-phosphates and 5'-hydroxyl ends, which occurs as a result of single-strand cleaving mechanism. As the name implies, it functions optimally at acid pH because it is commonly found in low pH environment of lysosomes. The action of DNase occurs in three phases. The initial phase introduces multiple nicks in the phosphodiester backbone. The second phase produces acid-soluble nucleotides. The third phase, which is the terminal phase, consists of hyperchromic shift resulting from reduction of oligonucleotides. There are several known DNases II, including: * DNase II alpha (usually known as DNase II), which is thought to be ubiquitously expressed in human tissue. It has been ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
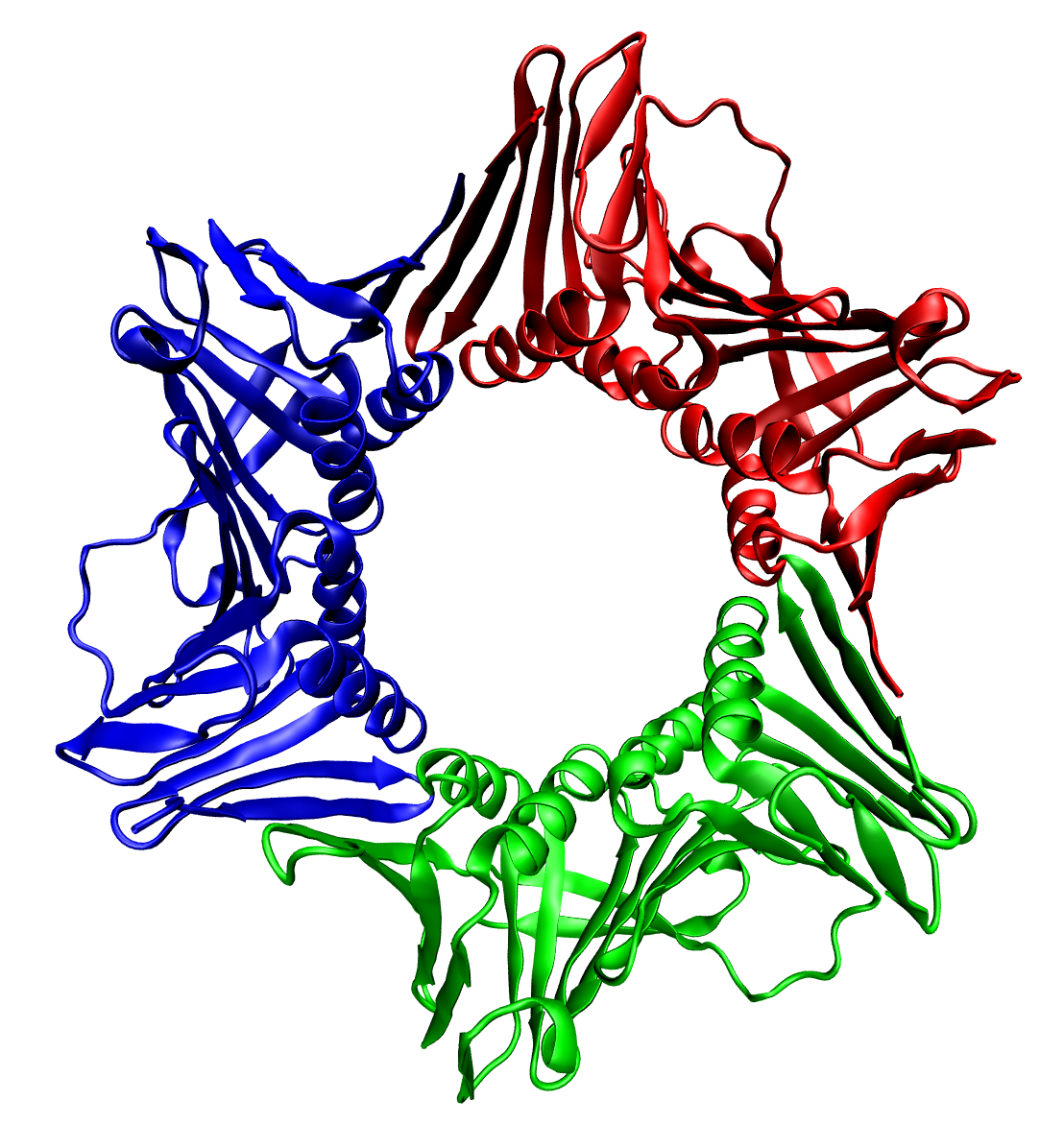
Protein Quaternary Structure
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also referred to as subunits). Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors. Description and examples Many proteins are actually assemblies of multiple polypeptide chains. The quaternary structure refers to the number and arrangement of the protein subunits w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dimer
In biochemistry, a protein dimer is a macromolecular complex or protein multimer, multimer formed by two protein monomers, or single proteins, which are usually Non-covalent interaction, non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' has roots meaning "two parts", ''wikt:di-#Prefix, di-'' + ''wikt:-mer#Suffix, -mer''. A protein dimer is a type of protein quaternary structure. A protein homodimer is formed by two identical proteins while a protein heterodimer is formed by two different proteins. Most protein dimers in biochemistry are not connected by covalent bonds. An example of a non-covalent heterodimer is the enzyme reverse transcriptase, which is composed of two different amino acid chains. An exception is dimers that are linked by disulfide bridges such as the homodimeric protein IKBKG, NEMO. Some proteins contain specialized domains to ensure dimerization (dimerization domains) and specificity. The G protein- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monomer
A monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or two- or three-dimensional network in a process called polymerization. Classification Chemistry classifies monomers by type, and two broad classes based on the type of polymer they form. By type: * natural vs synthetic, e.g. glycine vs caprolactam, respectively * polar vs nonpolar, e.g. vinyl acetate vs ethylene, respectively * cyclic vs linear, e.g. ethylene oxide vs ethylene glycol, respectively By type of polymer they form: * those that participate in condensation polymerization * those that participate in addition polymerization Differing stoichiometry causes each class to create its respective form of polymer. : The polymerization of one kind of monomer gives a homopolymer. Many polymers are copolymers, meaning that they are derived from two different monomers. In the case of condensation polymerizations, t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dimethyl Sulfoxide
Dimethyl sulfoxide (DMSO) is an organosulfur compound with the formula . This colorless liquid is the sulfoxide most widely used commercially. It is an important polar aprotic solvent that dissolves both polar and nonpolar compounds and is miscible in a wide range of organic solvents as well as water. It has a relatively high boiling point. DMSO is metabolised to compounds that leave a garlic-like taste in the mouth after DMSO is absorbed by skin. In terms of chemical structure, the molecule has idealized Cs symmetry. It has a trigonal pyramidal molecular geometry consistent with other three-coordinate S(IV) compounds, with a nonbonded electron pair on the approximately tetrahedral sulfur atom. Synthesis and production Dimethyl sulfoxide was first synthesized in 1866 by the Russian scientist Alexander Zaytsev, who reported his findings in 1867. Its modern use as an industrial solvent began through popularization by Thor Smedslund at the Stepan Chemical Company. Dimeth ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
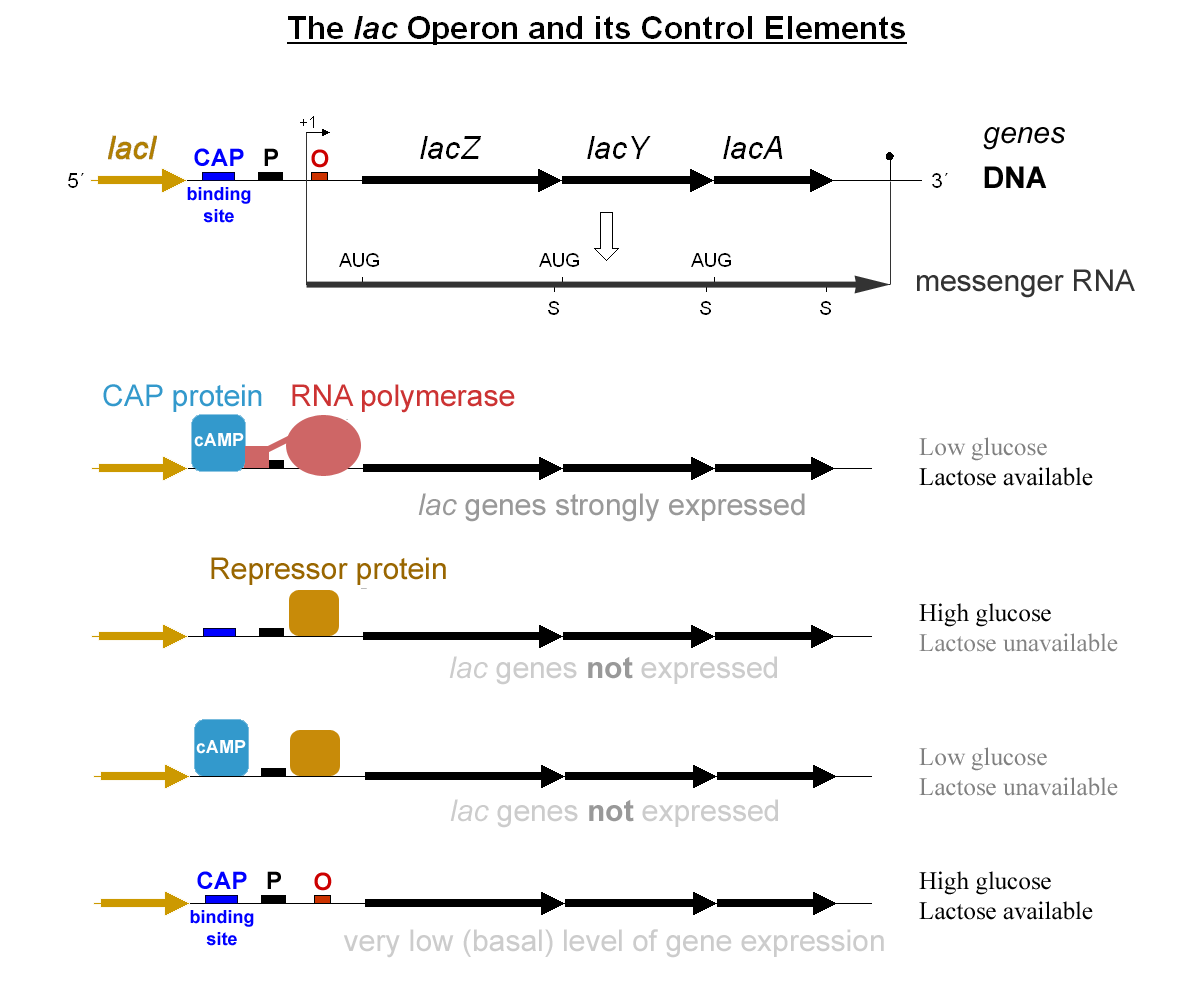
Activator Sequence
A transcriptional activator is a protein (transcription factor) that increases transcription of a gene or set of genes. Activators are considered to have ''positive'' control over gene expression, as they function to promote gene transcription and, in some cases, are required for the transcription of genes to occur. Most activators are DNA-binding proteins that bind to enhancers or promoter-proximal elements. The DNA site bound by the activator is referred to as an "activator-binding site". The part of the activator that makes protein–protein interactions with the general transcription machinery is referred to as an "activating region" or "activation domain". Most activators function by binding sequence-specifically to a regulatory DNA site located near a promoter and making protein–protein interactions with the general transcription machinery (RNA polymerase and general transcription factors), thereby facilitating the binding of the general transcription machinery to the pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |