|
Atg1
AuTophaGy related 1 (Atg1) is a 101.7kDa serine/threonine kinase in ''S.cerevisiae'', encoded by the gene ATG1. It is essential for the initial building of the autophagosome and Cvt vesicles. In a non-kinase role it is - through complex formation with Atg13 and Atg17 - directly controlled by the TOR kinase, a sensor for nutrient availability. Introduction Atg1 can associate with a number of other proteins of the Atg family to form a complex that functions in autophagosome or Cvt vesicle formation. The initiation of autophagy involves the building of the pre-autophagosomal structure (PAS). Most Atg proteins accumulate at the PAS and generate either Cvt vesicles under normal growing conditions or autophagosomes under starvation. To date, there are 31 ATG genes, which can be classified into several different groups according to their functions at the different steps of the pathway. 17 of these genes only work in the Cvt pathway. Structure The Atg1 gene lies on chromosome VII of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Atg1 Interaction Partners
AuTophaGy related 1 (Atg1) is a 101.7kDa serine/threonine kinase in ''S.cerevisiae'', encoded by the gene ATG1. It is essential for the initial building of the autophagosome and Cytoplasm-to-vacuole targeting, Cvt vesicles. In a non-kinase role it is - through complex formation with Atg13 and Atg17 - directly controlled by the TOR (gene), TOR kinase, a sensor for nutrient availability. Introduction Atg1 can associate with a number of other proteins of the Atg family to form a complex that functions in autophagosome or Cvt vesicle formation. The initiation of autophagy involves the building of the pre-autophagosomal structure (PAS). Most Atg proteins accumulate at the PAS and generate either Cvt vesicles under normal growing conditions or autophagosomes under starvation. To date, there are 31 ATG genes, which can be classified into several different groups according to their functions at the different steps of the pathway. 17 of these genes only work in the Cvt pathway. Structure ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
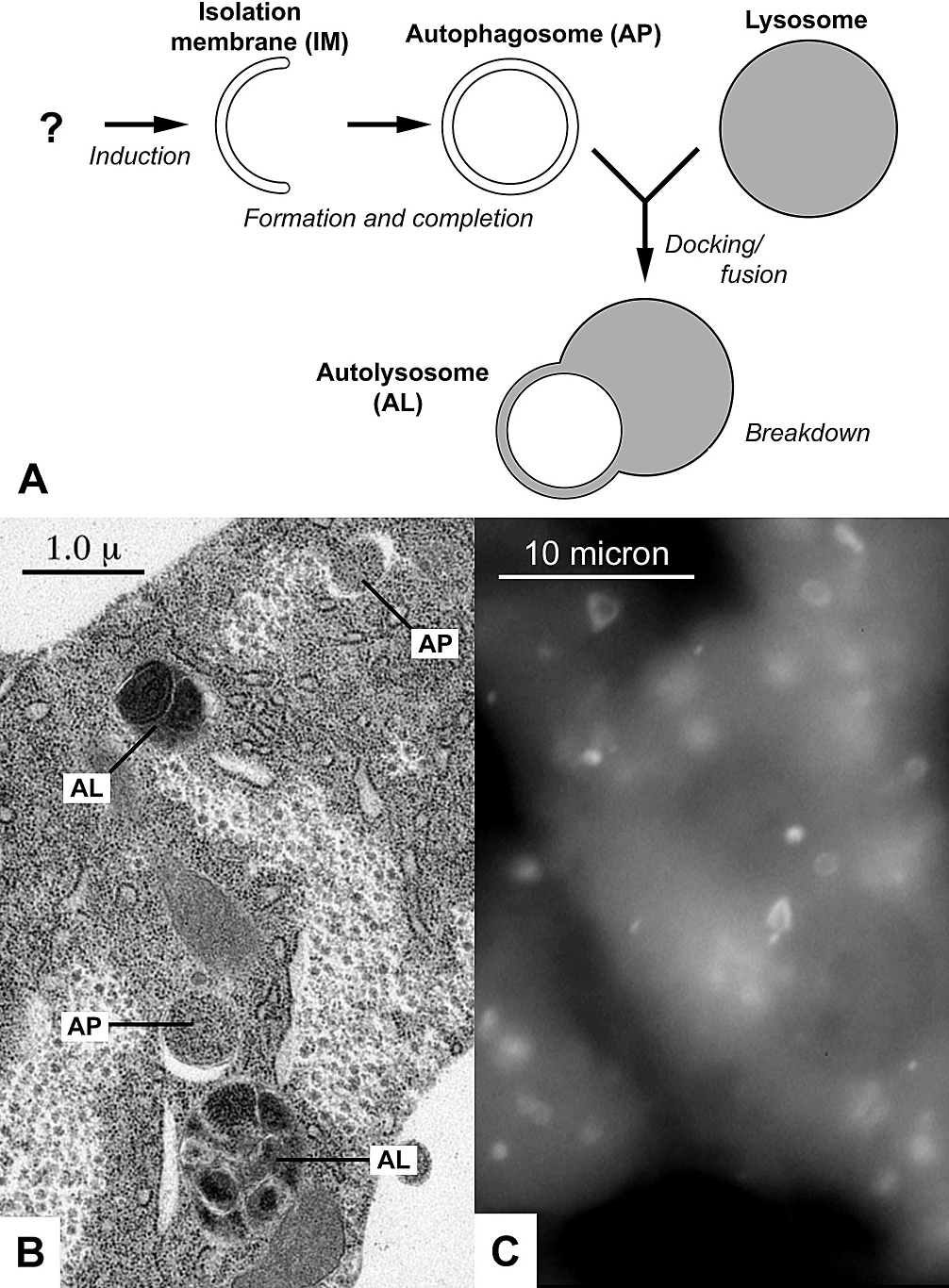
Autophagosome
An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents (e.g., abnormal intracellular proteins, excess or damaged organelles, invading microorganisms). After formation, autophagosomes deliver cytoplasmic components to the lysosomes. The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome's hydrolases degrade the autophagosome-delivered contents and its inner membrane. The formation of autophagosomes is regulated by genes that are well-conserved from yeast to higher eukaryotes. The nomenclature of these genes has differed from paper to paper, but it has been simplified in recent years. The gene families formerly known as APG, AUT, CVT, GSA, PAZ, and PDD are now unified as the ATG (AuTophaGy related) family. The size of autophagosomes vary between mammals and yeast. Yeast autophagosomes are about 500-900 nm, while ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophagy
Autophagy (or autophagocytosis; from the Ancient Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components (like ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TOR (gene)
The mammalian target of rapamycin (mTOR), also referred to as the mechanistic target of rapamycin, and sometimes called FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the ''MTOR'' gene. mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases. mTOR links with other proteins and serves as a core component of two distinct protein complexes, mTOR complex 1 and mTOR complex 2, which regulate different cellular processes. In particular, as a core component of both complexes, mTOR functions as a serine/threonine protein kinase that regulates cell growth, cell proliferation, cell motility, cell survival, protein synthesis, autophagy, and transcription. As a core component of mTORC2, mTOR also functions as a tyrosine protein kinase that promotes the activation of insulin receptors and insulin-like growth factor 1 receptors. mTORC2 has also been implicated in the control and mainten ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ULK1
ULK1 is an enzyme that in humans is encoded by the ''ULK1'' gene. Unc-51 like autophagy activating kinase (ULK1/2) are two similar isoforms of an enzyme that in humans are encoded by the ''ULK1/2'' genes. ">/sup> ">/sup> It is specifically a kinase that is involved with autophagy, particularly in response to amino acid withdrawal. Not many studies have been done comparing the two isoforms, but some differences have been recorded. Function Ulk1/2 is an important protein in autophagy for mammalian cells, and is homologous to ATG1 in yeast. It is part of the ULK1-complex, which is needed in early steps of autophagosome biogenesis. The ULK1 complex also consists of the FAK family kinase interacting protein of 200 kDa (FIP200 or RB1CC1) and the HORMA (Hop/Rev7/Mad2) domain-containing proteins ATG13 and ATG101. ULK1, specifically, appears to be the most essential for autophagy and is activated under conditions of nutrient deprivation by several upstream signals which is followed b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Death
Cell death is the event of a biological cell ceasing to carry out its functions. This may be the result of the natural process of old cells dying and being replaced by new ones, as in programmed cell death, or may result from factors such as diseases, localized injury, or the death of the organism of which the cells are part. Apoptosis or Type I cell-death, and autophagy or Type II cell-death are both forms of programmed cell death, while necrosis is a non-physiological process that occurs as a result of infection or injury. Programmed cell death Programmed cell death (PCD) is cell death mediated by an intracellular program. PCD is carried out in a regulated process, which usually confers advantage during an organism's life-cycle. For example, the differentiation of fingers and toes in a developing human embryo occurs because cells between the fingers apoptose; the result is that the digits separate. PCD serves fundamental functions during both plant and metazoa (multic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the wings of bats and birds, the arms of primates, the front flippers of whales and the forelegs of four-legged vertebrates like dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adapted to different purposes as the result of descent with modification from a common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In developmental biology, organs that developed in the embryo in the same manner and from similar origins, such as from matc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Kinase A
In cell biology, protein kinase A (PKA) is a family of enzymes whose activity is dependent on cellular levels of cyclic AMP (cAMP). PKA is also known as cAMP-dependent protein kinase (). PKA has several functions in the cell, including regulation of glycogen, sugar, and lipid metabolism. It should not be confused with 5'-AMP-activated protein kinase ( AMP-activated protein kinase). History Protein kinase A, more precisely known as adenosine 3',5'-monophosphate (cyclic AMP)-dependent protein kinase, abbreviated to PKA, was discovered by chemists Edmond H. Fischer and Edwin G. Krebs in 1968. They won the Nobel Prize in Physiology or Medicine in 1992 for their work on phosphorylation and dephosphorylation and how it relates to PKA activity. PKA is one of the most widely researched protein kinases, in part because of its uniqueness; out of 540 different protein kinase genes that make up the human kinome, only one other protein kinase, casein kinase 2, is known to exist in a p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mechanism Of Atg1 Complex Regulation By TOR
Mechanism may refer to: *Mechanism (engineering), rigid bodies connected by joints in order to accomplish a desired force and/or motion transmission *Mechanism (biology), explaining how a feature is created *Mechanism (philosophy), a theory that all natural phenomena can be explained by physical causes *Mechanism (sociology), a theory that all social phenomena can be explained by the existence of a deterministic mechanism * "The Mechanism", song by Disclosure * ''The Mechanism'' (TV series), a Netflix TV series See also * Machine *Machine (mechanical) *Linkage (mechanical) *Mechanism design, the art of designing rules of a game to achieve a specific outcome *Mechanism of action, the means by which a drug exerts its biological effects *Defence mechanism, unconscious mechanisms aimed at reducing anxiety *Reaction mechanism, the sequence of reactions by which overall chemical change occurs *Antikythera mechanism, an ancient Greek analog computer *Theory of operation A theory of o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Saccharomyces Cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been originally isolated from the skin of grapes. It is one of the most intensively studied eukaryotic model organisms in molecular and cell biology, much like ''Escherichia coli'' as the model bacterium. It is the microorganism behind the most common type of fermentation. ''S. cerevisiae'' cells are round to ovoid, 5–10 μm in diameter. It reproduces by budding. Many proteins important in human biology were first discovered by studying their homologs in yeast; these proteins include cell cycle proteins, signaling proteins, and protein-processing enzymes. ''S. cerevisiae'' is currently the only yeast cell known to have Berkeley bodies present, which are involved in particular secretory pathways. Antibodies against ''S. cerevis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulation Of Atg1 Complex By TOR
Regulation is the management of complex systems according to a set of rules and trends. In systems theory, these types of rules exist in various fields of biology and society, but the term has slightly different meanings according to context. For example: * in biology, gene regulation and metabolic regulation allow living organisms to adapt to their environment and maintain homeostasis; * in government, typically regulation means stipulations of the delegated legislation which is drafted by subject-matter experts to enforce primary legislation; * in business, industry self-regulation occurs through self-regulatory organizations and trade associations which allow industries to set and enforce rules with less government involvement; and, * in psychology, self-regulation theory is the study of how individuals regulate their thoughts and behaviors to reach goals. Social Regulation in the social, political, psychological, and economic domains can take many forms: legal restriction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast Two Hybrid
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively. The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the DNA-binding domain (DBD or often also abbreviated as BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay. History Pioneered by Stanley Fields and Ok-Kyu Song in 1989, the technique was originally designed to detect protein–protei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |