|
PROSESS Input Entry Page
Protein Structure Evaluation Suite & Server (PROSESS) is a freely available web server for protein structure validation. It has been designed at the University of Alberta to assist with the process of evaluating and validating protein structures solved by NMR spectroscopy. Structure validation Structure validation is a particularly important component of the structure determination pipeline as many protein structures have small structural errors (i.e. distorted bond lengths or angles, incompatible torsion angles, overlapping atoms) that are not easily detected by visual inspection. For protein structures solved by NMR spectroscopy, where large numbers of structures are generated and where coordinate inaccuracies are common, this problem is particularly acute. Methodology Most NMR-based structure validation protocols primarily use NOE ( Nuclear Overhauser Enhancement), J-coupling or residual dipolar coupling ( RDC ) data to assess or validate structures. In particular, they ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Shift
In nuclear magnetic resonance (NMR) spectroscopy, the chemical shift is the resonant frequency of an atomic nucleus relative to a standard in a magnetic field. Often the position and number of chemical shifts are diagnostic of the structure of a molecule. Chemical shifts are also used to describe signals in other forms of spectroscopy such as photoemission spectroscopy. Some atomic nuclei possess a magnetic moment (nuclear spin), which gives rise to different energy levels and resonance frequencies in a magnetic field. The total magnetic field experienced by a nucleus includes local magnetic fields induced by currents of electrons in the molecular orbitals (electrons have a magnetic moment themselves). The electron distribution of the same type of nucleus (e.g. ) usually varies according to the local geometry (binding partners, bond lengths, angles between bonds, and so on), and with it the local magnetic field at each nucleus. This is reflected in the spin energy levels (an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DSSP (algorithm)
The DSSP algorithm is the standard method for assigning secondary structure to the amino acids of a protein, given the atomic-resolution coordinates of the protein. The abbreviation is only mentioned once in the 1983 paper describing this algorithm, where it is the name of the Pascal program that implements the algorithm ''Define Secondary Structure of Proteins''. Algorithm DSSP begins by identifying the intra-backbone hydrogen bonds of the protein using a purely electrostatic definition, assuming partial charges of −0.42 ''e'' and +0.20 ''e'' to the carbonyl oxygen and amide hydrogen respectively, their opposites assigned to the carbonyl carbon and amide nitrogen. A hydrogen bond is identified if ''E'' in the following equation is less than -0.5 kcal/mol: : E = 0.084 \left\ \cdot 332 \, \mathrm where the r_ terms indicate the distance between atoms A and B, taken from the carbon (C) and oxygen (O) atoms of the C=O group and the nitrogen (N) and hydrogen (H) atoms of the N-H ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Bioinformatics
Structural bioinformatics is the branch of bioinformatics that is related to the analysis and prediction of the three-dimensional structure of biological macromolecules such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs, principles of molecular folding, evolution, binding interactions, and structure/function relationships, working both from experimentally solved structures and from computational models. The term ''structural'' has the same meaning as in structural biology, and structural bioinformatics can be seen as a part of computational structural biology. The main objective of structural bioinformatics is the creation of new methods of analysing and manipulating biological macromolecular data in order to solve problems in biology and generate new knowledge. Introduction Protein structure The structure of a protein is directly related to its function. The presence of certa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Structure Prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure from Protein primary structure, primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology and addresses Levinthal's paradox. Accurate structure prediction has important applications in medicine (for example, in drug design) and biotechnology (for example, in novel enzyme design). Starting in 1994, the performance of current methods is assessed biannually in the ''Critical Assessment of Structure Prediction'' (CASP) experiment. A continuous evaluation of protein structure prediction web servers is performed by the community project ''Continuous Automated Model EvaluatiOn'' (CAMEO3D). Protein structure a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
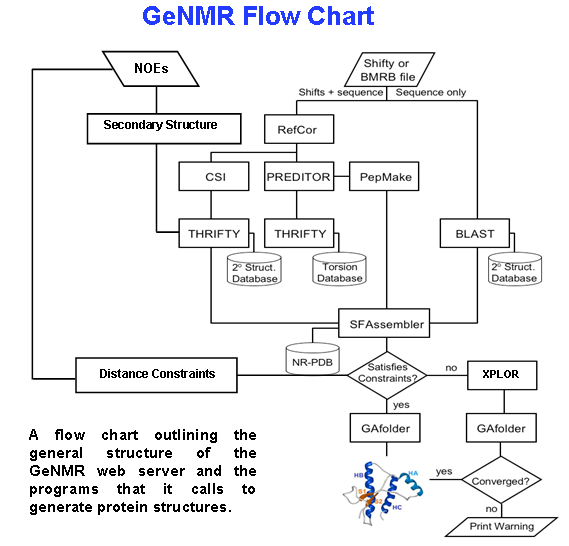
GeNMR
GeNMR method (GEnerate NMR structures) is the first fully automated template-based method of protein structure determination that utilizes both NMR chemical shifts and NOE-based distance restraints. In addition to the template-based approach, the GeNMR webserver also offers an ''ab initio'' protein folding mode that starts folding from an extended structure. The GeNMR web server produces an ensemble of PDB coordinates within a period ranging from 20 minutes to 4 hours, depending on protein size, server load, quality and type of experimental information, and selected protocol options. GeNMR webserver is composed of two parts, a front-end web-interface (written in Perl and HTML) and a back-end consisting of eight different alignment, structure generation and structure optimization programs along with three local databases. Input GeNMR accepts and processes backbone and side chain 1H, 13C or 15N chemical shift data of almost any combination (HA only, HN only, HA+HN only, HA+HN+si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Random Coil Index
Random coil index (RCI) predicts protein flexibility by calculating an inverse weighted average of backbone secondary chemical shifts and predicting values of model-free order parameters as well as per-residue RMSD of NMR and molecular dynamics ensembles from this parameter. The key advantages of this protocol over existing methods of studying protein flexibility are # it does not require prior knowledge of a protein's tertiary structure, # it is not sensitive to the protein's overall tumbling and # it does not require additional NMR measurements beyond the standard experiments for backbone assignments. The application of secondary chemical shifts to characterize protein flexibility is based on an assumption that the proximity of chemical shifts to random coil values is a manifestation of increased protein mobility, while significant differences from random coil values are an indication of a relatively rigid structure. Even though chemical shifts of rigid residues may adopt ran ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Dynamics
In molecular biology, proteins are generally thought to adopt unique structures determined by their amino acid sequences. However, proteins are not strictly static objects, but rather populate ensembles of (sometimes similar) conformations. Transitions between these states occur on a variety of length scales (tenths of angstroms to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. The study of protein dynamics is most directly concerned with the transitions between these states, but can also involve the nature and equilibrium populations of the states themselves. These two perspectives— kinetics and thermodynamics, respectively—can be conceptually synthesized in an " energy landscape" paradigm: highly populated states and the kinetics of transitions between them can be described by the depths of energy wells and the heights of energy barriers, respectively. Local flexibility: atoms ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Nuclear Magnetic Resonance Spectroscopy
Nuclear magnetic resonance spectroscopy of proteins (usually abbreviated protein NMR) is a field of structural biology in which NMR spectroscopy is used to obtain information about the structure and dynamics of proteins, and also nucleic acids, and their complexes. The field was pioneered by Richard R. Ernst and Kurt Wüthrich at the ETH, and by Ad Bax, Marius Clore, Angela Gronenborn at the NIH, and Gerhard Wagner at Harvard University, among others. Structure determination by NMR spectroscopy usually consists of several phases, each using a separate set of highly specialized techniques. The sample is prepared, measurements are made, interpretive approaches are applied, and a structure is calculated and validated. NMR involves the quantum-mechanical properties of the central core ("nucleus") of the atom. These properties depend on the local molecular environment, and their measurement provides a map of how the atoms are linked chemically, how close they are in space, and how ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Magnetic Resonance Spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a Spectroscopy, spectroscopic technique based on re-orientation of Atomic nucleus, atomic nuclei with non-zero nuclear spins in an external magnetic field. This re-orientation occurs with absorption of electromagnetic radiation in the radio frequency region from roughly 4 to 900 MHz, which depends on the Isotope, isotopic nature of the nucleus and increases proportionally to the strength of the external magnetic field. Notably, the resonance frequency of each NMR-active nucleus depends on its chemical environment. As a result, NMR spectra provide information about individual functional groups present in the sample, as well as about connections between nearby nuclei in the same molecule. As the NMR spectra are unique or highly characteristic to individual compounds and functional groups, NMR spectroscopy is one of the most important methods to identify ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
VADAR
Volume, Area, Dihedral Angle Reporter (VADAR) is a freely available protein structure validation web server that was developed as a collaboration between Dr. Brian Sykes and Dr. David Wishart at the University of Alberta. VADAR consists of over 15 different algorithms and programs for assessing and validating peptide and protein structures from their PDB coordinate data. VADAR is capable of determining secondary structure (using three different algorithms), identifying and classifying six different types of beta turns, determining and calculating the strength of C=O -- N-H hydrogen bonds, calculating residue-specific accessible surface areas (ASA), calculating residue volumes, determining backbone and side chain torsion angles (phi, psi, omega and chi angles), assessing local structure quality (through numerous quality indices), evaluating global structure quality, and identifying residue "outliers" (residues with unusual structural features). The results have been validated th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |