|
Endonuclease V
Endonuclease V ''(endoV)'' is a highly conserved endonuclease enzyme family. The primary function of endoV differs significantly in prokaryotes and eukaryotes, as suggested by studies on the ''E. coli'' and human orthologs. In prokaryotes endoV is primarily a deoxyribonuclease involved in DNA repair of deoxyinosine introduced into the genome by deamidation of adenine bases ( EC 3.1.21.7). However, it has broad substrate specificity and can also act on other types of DNA lesions as well as on inosine-containing RNA. In eukaryotes endoV is primarily a ribonuclease and cleaves single-stranded RNA at the 3' position relative to an inosine base, which may be present due to RNA editing by deaminase enzymes (EC 3.1.26.-). The human endoV localizes to the cytoplasm and nucleoli, suggesting a possible role in processes involving ribosomal RNA Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Crystallography
Crystallography is the branch of science devoted to the study of molecular and crystalline structure and properties. The word ''crystallography'' is derived from the Ancient Greek word (; "clear ice, rock-crystal"), and (; "to write"). In July 2012, the United Nations recognised the importance of the science of crystallography by proclaiming 2014 the International Year of Crystallography.UN announcement "International Year of Crystallography" iycr2014.org. 12 July 2012 Crystallography is a broad topic, and many of its subareas, such as X-ray crystallography, are themselves important scientific topics. Crystallography ranges from the fundamentals of crystal structure to the mathematics of Crystal system, crystal geometry, including those that are Aperiodic crystal, not periodic or quasi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is complementary and pairs to either thymine in DNA or uracil in RNA. In cells adenine, as an independent molecule, is rare. It is almost always covalent bond, covalently bound to become a part of a larger biomolecule. Adenine has a central role in cellular respiration. It is part of adenosine triphosphate which provides the energy that drives and supports most activities in living cell (biology), cells, such as Protein biosynthesis, protein synthesis, chemical synthesis, muscle contraction, and nerve impulse propagation. In respiration it also participates as part of the cofactor (biochemistry), cofactors nicotinamide adenine dinucleotide, flavin adenine dinucleotide, and Coenzyme A. It is also part of adenosine, adenosine monophosphate, cy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form SSU rRNA, small and LSU rRNA, large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and Translation (biology), translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins, though this ratio differs between Prokaryote, prokaryotes and Eukaryote, eukaryotes. Structure Although the primary structure of rRNA sequences can vary across organisms, Base pair, base-pairing within these sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
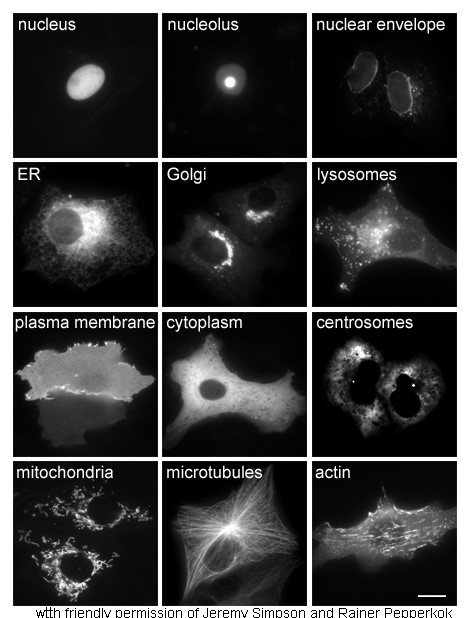
Nucleoli
The nucleolus (; : nucleoli ) is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of the nucleolus is the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy. History The nucleolus was identified by bright-field microscopy during the 1830s. Theodor Schwann in his 1839 treatise described that Schleiden had identified small corpuscles in nuclei, and named the structures "Kernkörperchen". In a 1947 translation of the work to English, the structure was named "nucleolus". Little was known about the function of the nucleolus until 1964, when a study of nucleoli by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell and contained within the nuclear membrane is termed the nucleoplasm. The main components of the cytoplasm are the cytosol (a gel-like substance), the cell's internal sub-structures, and various cytoplasmic inclusions. In eukaryotes the cytoplasm also includes the nucleus, and other membrane-bound organelles.The cytoplasm is about 80% water and is usually colorless. The submicroscopic ground cell substance, or cytoplasmic matrix, that remains after the exclusion of the cell organelles and particles is groundplasm. It is the hyaloplasm of light microscopy, a highly complex, polyphasic system in which all resolvable cytoplasmic elements are suspended, including the larger organelles such as the ribosomes, mitochondria, plant plasti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deaminase
Deamination is the removal of an amino group from a molecule. Enzymes that catalyse this reaction are called deaminases. In the human body, deamination takes place primarily in the liver; however, it can also occur in the kidney. In situations of excess protein intake, deamination is used to break down amino acids for energy. The amino group is removed from the amino acid and converted to ammonia. The rest of the amino acid is made up of mostly carbon and hydrogen, and is recycled or oxidized for energy. Ammonia is toxic to the human system, and enzymes convert it to urea or uric acid by addition of carbon dioxide molecules (which is not considered a deamination process) in the urea cycle, which also takes place in the liver. Urea and uric acid can safely diffuse into the blood and then be excreted in urine. Deamination reactions in DNA Cytosine Spontaneous deamination is the hydrolysis reaction of cytosine into uracil, releasing ammonia in the process. This can occur in vitro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Editing
RNA editing (also RNA modification) is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing (e.g. RNA splicing, splicing, 5'-capping enzyme, capping, and 3'-polyadenylation) not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases. RNA editing has been observed in some tRNA, rRNA, mRNA, or microRNA, miRNA molecules of eukaryotes and their viruses, archaea, and prokaryotes. RNA editing occurs in the cell nucleus, as well as within mitochondria and plastids. In vertebrates, editing is rare and usually ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single-stranded
When referring to DNA transcription, the coding strand (or informational strand) is the DNA strand whose base sequence is identical to the base sequence of the RNA transcript produced (although with thymine replaced by uracil). It is this strand which contains codons, while the non-coding strand contains anticodons. During transcription, RNA Pol II binds to the non-coding template strand, reads the anti-codons, and transcribes their sequence to synthesize an RNA transcript with complementary bases. By convention, the coding strand is the strand used when displaying a DNA sequence. It is presented in the 5' to 3' direction. Wherever a gene exists on a DNA molecule, one strand is the coding strand (or sense strand), and the other is the noncoding strand (also called the antisense strand, anticoding strand, template strand or transcribed strand). Strands in transcription bubble During transcription, RNA polymerase unwinds a short section of the DNA double helix near the s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Inosine
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring (also known as a ribofuranose) via a β-N9-glycosidic bond. It was discovered in 1965 in analysis of RNA transferase. Inosine is commonly found in tRNAs and is essential for proper translation of the genetic code in wobble base pairs. Knowledge of inosine metabolism has led to advances in immunotherapy in recent decades. Inosine monophosphate is oxidised by the enzyme inosine monophosphate dehydrogenase, yielding xanthosine monophosphate, a key precursor in purine metabolism. Mycophenolate mofetil is an anti-metabolite, anti-proliferative drug that acts as an inhibitor of inosine monophosphate dehydrogenase. It is used in the treatment of a variety of autoimmune diseases including granulomatosis with polyangiitis because the uptake of purine by actively dividing B cells can exceed 8 times that of normal body cells, and, therefore, this set of white cells (which cannot operate purine salvage ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in cells, by internal metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair processes fail, including apoptosis, irreparable DNA damage may occur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substrate (chemistry)
In chemistry, the term substrate is highly context-dependent. Broadly speaking, it can refer either to a chemical species being observed in a chemical reaction, or to a surface on which other chemical reactions or microscopy are performed. In the former sense, a reagent is added to the ''substrate'' to generate a product through a chemical reaction. The term is used in a similar sense in synthetic and organic chemistry, where the substrate is the chemical of interest that is being modified. In biochemistry, an enzyme substrate is the material upon which an enzyme acts. When referring to Le Chatelier's principle, the substrate is the reagent whose concentration is changed. ;Spontaneous reaction : :*Where S is substrate and P is product. ;Catalysed reaction : :*Where S is substrate, P is product and C is catalyst. In the latter sense, it may refer to a surface on which other chemical reactions are performed or play a supporting role in a variety of spectroscopic and micr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |