Transposons As A Genetic Tool on:
[Wikipedia]
[Google]
[Amazon]
 A transposable element (TE), also transposon, or jumping gene, is a type of
A transposable element (TE), also transposon, or jumping gene, is a type of
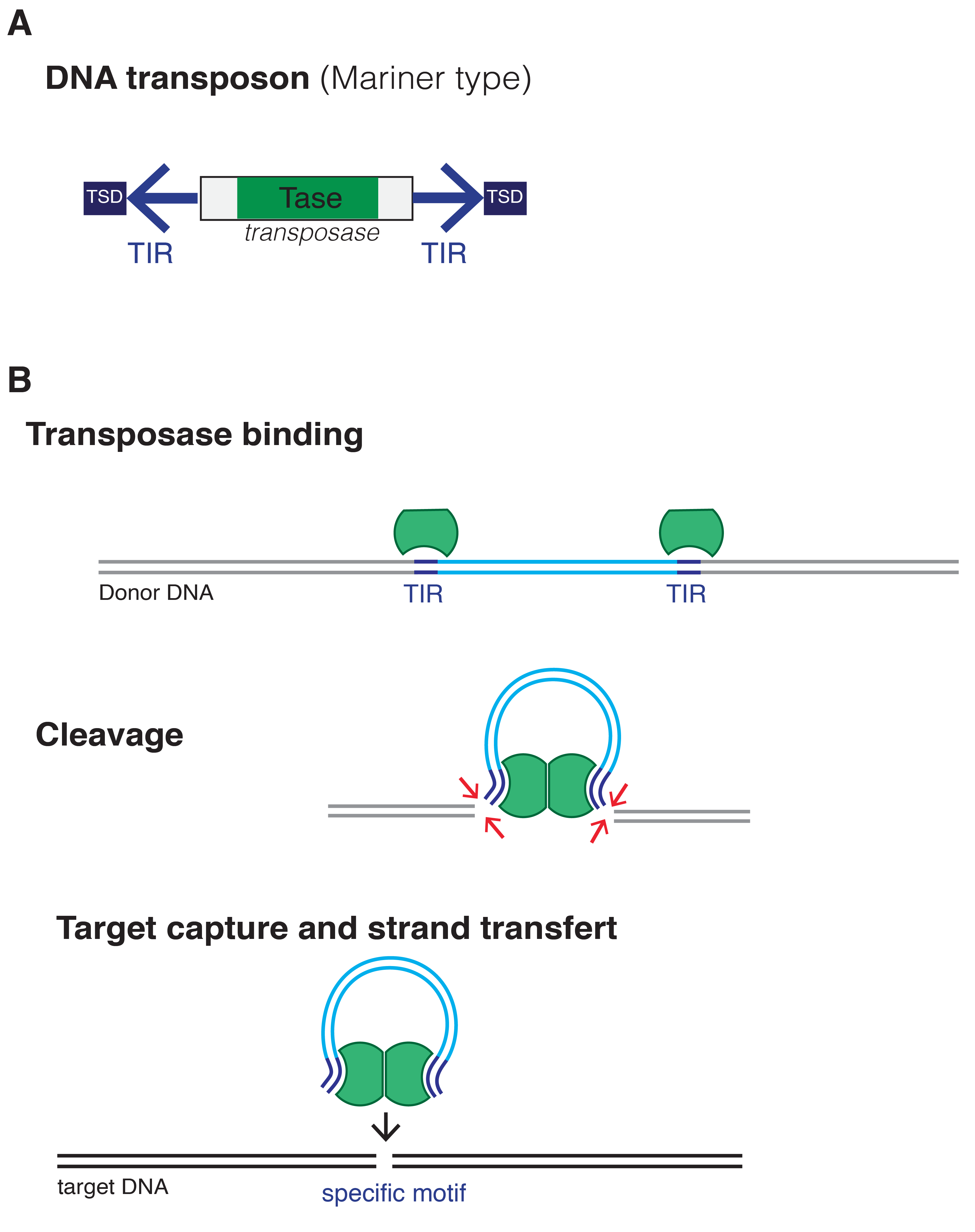 The cut-and-paste transposition mechanism of class II TEs does not involve an RNA intermediate. The transpositions are catalyzed by several
The cut-and-paste transposition mechanism of class II TEs does not involve an RNA intermediate. The transpositions are catalyzed by several
Repbase
– a database of transposable element sequences
Dfam
- a database of transposable element families, multiple sequence alignments, and sequence models
RepeatMasker
– a computer program used by computational biologists to
Use of the Sleeping Beauty Transposon System for Stable Gene Expression in Mouse Embryonic Stem Cells
Introduction to Transposons, 2018 YouTube video
{{Authority control Modification of genetic information Mobile genetic elements Molecular biology Non-coding DNA
mobile genetic element
Mobile genetic elements (MGEs), sometimes called selfish genetic elements, are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In ...
, a nucleic acid sequence in DNA that can change its position within a genome.
The discovery of mobile genetic elements earned Barbara McClintock
Barbara McClintock (June 16, 1902 – September 2, 1992) was an American scientist and cytogenetics, cytogeneticist who was awarded the 1983 Nobel Prize in Physiology or Medicine. McClintock received her PhD in botany from Cornell University ...
a Nobel Prize in 1983.
There are at least two classes of TEs: Class I TEs or retrotransposon
Retrotransposons (also called Class I transposable elements) are mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or ...
s generally function via reverse transcription
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B virus, hepatitis B to replicate their genomes, by retrot ...
, while Class II TEs or DNA transposon DNA transposons are DNA sequences, sometimes referred to "jumping genes", that can move and integrate to different locations within the genome. They are class II transposable elements (TEs) that move through a DNA intermediate, as opposed to class I ...
s encode the protein transposase
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transpositio ...
, which they require for insertion and excision, and some of these TEs also encode other proteins.
Discovery by Barbara McClintock
Barbara McClintock
Barbara McClintock (June 16, 1902 – September 2, 1992) was an American scientist and cytogenetics, cytogeneticist who was awarded the 1983 Nobel Prize in Physiology or Medicine. McClintock received her PhD in botany from Cornell University ...
discovered the first TEs in maize
Maize (; ''Zea mays''), also known as corn in North American English, is a tall stout grass that produces cereal grain. It was domesticated by indigenous peoples in southern Mexico about 9,000 years ago from wild teosinte. Native American ...
(''Zea mays'') at the Cold Spring Harbor Laboratory
Cold Spring Harbor Laboratory (CSHL) is a private, non-profit institution with research programs focusing on cancer, neuroscience, botany, genomics, and quantitative biology. It is located in Laurel Hollow, New York, in Nassau County, on ...
in New York. McClintock was experimenting with maize plants that had broken chromosomes.
In the winter of 1944–1945, McClintock planted corn kernels that were self-pollinated, meaning that the silk (style
Style, or styles may refer to:
Film and television
* ''Style'' (2001 film), a Hindi film starring Sharman Joshi, Riya Sen, Sahil Khan and Shilpi Mudgal
* ''Style'' (2002 film), a Tamil drama film
* ''Style'' (2004 film), a Burmese film
* '' ...
) of the flower received pollen from its own anther
The stamen (: stamina or stamens) is a part consisting of the male reproductive organs of a flower. Collectively, the stamens form the androecium., p. 10
Morphology and terminology
A stamen typically consists of a stalk called the filament ...
. These kernels came from a long line of plants that had been self-pollinated, causing broken arms on the end of their ninth chromosomes. As the maize plants began to grow, McClintock noted unusual color patterns on the leaves. For example, one leaf had two albino patches of almost identical size, located side by side on the leaf. McClintock hypothesized that during cell division certain cells lost genetic material, while others gained what they had lost. However, when comparing the chromosomes of the current generation of plants with the parent generation, she found certain parts of the chromosome had switched position. This refuted the popular genetic theory of the time that genes were fixed in their position on a chromosome. McClintock found that genes could not only move but they could also be turned on or off due to certain environmental conditions or during different stages of cell development.
McClintock also showed that gene mutations could be reversed. She presented her report on her findings in 1951, and published an article on her discoveries in ''Genetics'' in November 1953 entitled "Induction of Instability at Selected Loci in Maize".
At the 1951 Cold Spring Harbor Symposium where she first publicized her findings, her talk was met with silence. Her work was largely dismissed and ignored until the late 1960s–1970s when, after TEs were found in bacteria, it was rediscovered. She was awarded a Nobel Prize in Physiology or Medicine
The Nobel Prize in Physiology or Medicine () is awarded yearly by the Nobel Assembly at the Karolinska Institute for outstanding discoveries in physiology or medicine. The Nobel Prize is not a single prize, but five separate prizes that, acco ...
in 1983 for her discovery of TEs, more than thirty years after her initial research.
Classification
Transposable elements represent one of several types ofmobile genetic elements
Mobile genetic elements (MGEs), sometimes called selfish genetic elements, are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In ...
. TEs are assigned to one of two classes according to their mechanism of transposition, which can be described as either ''copy and paste'' (Class I TEs) or ''cut and paste'' (Class II TEs).
Retrotransposon
Class I TEs are copied in two stages: first, they are transcribed from DNA toRNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
, and the RNA produced is then reverse transcribed
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobile g ...
to DNA. This copied DNA is then inserted back into the genome at a new position. The reverse transcription step is catalyzed by a reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobi ...
, which is often encoded by the TE itself. The characteristics of retrotransposons are similar to retrovirus
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase e ...
es, such as HIV
The human immunodeficiency viruses (HIV) are two species of '' Lentivirus'' (a subgroup of retrovirus) that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the im ...
.
Despite the potential negative effects of retrotransposons, like inserting itself into the middle of a necessary DNA sequence, which can render important genes unusable, they are still essential to keep a species' ribosomal DNA
The ribosomal DNA (rDNA) consists of a group of ribosomal RNA encoding genes and related regulatory elements, and is widespread in similar configuration in all domains of life. The ribosomal DNA encodes the non-coding ribosomal RNA, integral struc ...
intact over the generations, preventing infertility.
Retrotransposons are commonly grouped into three main orders:
* Retrotransposons, with long terminal repeat
A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus o ...
s (LTRs), which encode reverse transcriptase, similar to retroviruses
* Retroposons, long interspersed nuclear elements (LINEs, LINE-1s, or L1s), which encode reverse transcriptase but lack LTRs, and are transcribed by RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ...
* Short interspersed nuclear element
Short interspersed nuclear elements (SINEs) are non-autonomous, non-coding transposable elements (TEs) that are about 100 to 700 base pairs in length. They are a class of retrotransposons, DNA elements that amplify themselves throughout eukaryo ...
s (SINEs) do not encode reverse transcriptase and are transcribed by RNA polymerase III
In eukaryote cells, RNA polymerase III (also called Pol III) is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA, and other small RNAs.
The genes transcribed by RNA Pol III fall in the category of "housekeeping" genes whose ex ...
Retroviruses can also be considered TEs. For example, after the conversion of retroviral RNA into DNA inside a host
A host is a person responsible for guests at an event or for providing hospitality during it.
Host may also refer to:
Places
* Host, Pennsylvania, a village in Berks County
* Host Island, in the Wilhelm Archipelago, Antarctica
People
* ...
cell, the newly produced retroviral DNA is integrated into the genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
of the host cell. These integrated DNAs are termed ''provirus
A provirus is a virus genome that is integrated into the DNA of a host cell. In the case of bacterial viruses (bacteriophages), proviruses are often referred to as prophages. However, proviruses are distinctly different from prophages and these te ...
es''. The provirus is a specialized form of eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
retrotransposon, which can produce RNA intermediates that may leave the host cell and infect other cells. The transposition cycle of retroviruses has similarities to that of prokaryotic
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
TEs, suggesting a distant relationship between the two.
DNA transposons
 The cut-and-paste transposition mechanism of class II TEs does not involve an RNA intermediate. The transpositions are catalyzed by several
The cut-and-paste transposition mechanism of class II TEs does not involve an RNA intermediate. The transpositions are catalyzed by several transposase
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transpositio ...
enzymes. Some transposases non-specifically bind to any target site in DNA, whereas others bind to specific target sequences. The transposase makes a staggered cut at the target site producing sticky ends
DNA ends refer to the properties of the ends of linear DNA molecules, which in molecular biology are described as "sticky" or "blunt" based on the shape of the complementary strands at the terminus. In sticky ends, one strand is longer than the o ...
, cuts out the DNA transposon and ligates it into the target site. A DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
fills in the resulting gaps from the sticky ends and DNA ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such ...
closes the sugar-phosphate backbone. This results in target site duplication and the insertion sites of DNA transposons may be identified by short direct repeats (a staggered cut in the target DNA filled by DNA polymerase) followed by inverted repeat
An inverted repeat (or IR) is a single stranded sequence of nucleotides followed downstream by its complementarity (molecular biology), reverse complement. The intervening sequence of nucleotides between the initial sequence and the reverse complem ...
s (which are important for the TE excision by transposase
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transpositio ...
).
Cut-and-paste TEs may be duplicated if their transposition takes place during S phase
S phase (Synthesis phase) is the phase of the cell cycle in which DNA is replicated, occurring between G1 phase and G2 phase. Since accurate duplication of the genome is critical to successful cell division, the processes that occur during S ...
of the cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
, when a donor site has already been replicated but a target site has not yet been replicated. Such duplications at the target site can result in gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene ...
, which plays an important role in genomic evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
.
Not all DNA transposons transpose through the cut-and-paste mechanism. In some cases, a replicative transposition Replicative transposition is a mechanism of transposition in molecular biology, proposed by James A. Shapiro in 1979, in which the transposable element is duplicated during the reaction, so that the transposing entity is a copy of the original elem ...
is observed in which a transposon replicates itself to a new target site (e.g. helitron).
Class II TEs comprise less than 2% of the human genome, making the rest Class I.
Autonomous and non-autonomous
Transposition can be classified as either "autonomous" or "non-autonomous" in both Class I and Class II TEs. Autonomous TEs can move by themselves, whereas non-autonomous TEs require the presence of another TE to move. This is often because dependent TEs lack transposase (for Class II) or reverse transcriptase (for Class I). Activator element (''Ac'') is an example of an autonomous TE, and dissociation elements (''Ds'') is an example of a non-autonomous TE. Without ''Ac,'' ''Ds'' is not able to transpose.Class III
Some researchers also identify a third class of transposable elements, which has been described as "a grab-bag consisting of transposons that don't clearly fit into the other two categories". Examples of such TEs are the Foldback (FB) elements of ''Drosophila melanogaster'', the TU elements of ''Strongylocentrotus purpuratus
''Strongylocentrotus purpuratus'' is a species of sea urchin in the family Strongylocentrotidae commonly known as the purple sea urchin. It lives along the eastern edge of the Pacific Ocean extending from Ensenada, Mexico, to British Columbi ...
'', and Miniature Inverted-repeat Transposable Elements Miniature Inverted-repeat Transposable Elements (MITEs) are a group of non-autonomous Class II transposable elements (DNA sequences). Being non-autonomous, MITEs cannot code for their own transposase. They exist within the genomes of animals, plant ...
.
Negative effects
Transposons can damage the genome of their host cell in different ways: * A transposon can insert into a functional gene and disable that gene. * After a DNA transposon is excised, the resulting gap may not be repaired correctly. * Many TEs contain promoters that drive transcription of their own genes. These promoters can cause aberrant expression of linked genes.Diseases
Diseases often caused by TEs include *Hemophilia
Haemophilia (British English), or hemophilia (American English) (), is a mostly inherited genetic disorder that impairs the body's ability to make blood clots, a process needed to stop bleeding. This results in people bleeding for a long ...
A and B
** LINE1
LINE1 (an abbreviation of Long interspersed nuclear element-1, also known as L1 and LINE-1) is a family of related class I transposable elements in the DNA of many groups of eukaryotes, including animals and plants, classified with the long in ...
(L1) TEs that land on the human Factor VIII have been shown to cause haemophiliaKazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE (March 1988). "Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man". ''Nature''. 332 (6160): 164–6. Bibcode
The bibcode (also known as the refcode) is a compact identifier used by several astronomical data systems to uniquely specify literature references.
Adoption
The Bibliographic Reference Code (refcode) was originally developed to be used in SIM ...
:1988Natur.332..164K. doi:10.1038/332164a0. PMID
PubMed is an openly accessible, free database which includes primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine (NLM) at the National Institutes of ...
2831458.
* Severe combined immunodeficiency
Severe combined immunodeficiency (SCID), also known as Swiss-type agammaglobulinemia, is a rare genetic disorder characterized by the disturbed development of functional T cells and B cells caused by numerous genetic mutations that result in diff ...
** Insertion of L1 into the APC gene causes colon cancer, confirming that TEs play an important role in disease development.
* Porphyria
Porphyria ( or ) is a group of disorders in which substances called porphyrins build up in the body, adversely affecting the skin or nervous system. The types that affect the nervous system are also known as Porphyria#Acute porphyrias, acute p ...
**Insertion of Alu element
An Alu element is a short stretch of DNA originally characterized by the action of the ''Arthrobacter luteus (Alu)'' restriction endonuclease. ''Alu'' elements are the most abundant transposable elements in the human genome, present in excess o ...
into the PBGD gene leads to interference with the coding region and leads to acute intermittent porphyria (AIP).
* Predisposition to cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
**LINE1(L1) TE's and other retrotransposons have been linked to cancer because they cause genomic instability.
* Duchenne muscular dystrophy
Duchenne muscular dystrophy (DMD) is a severe type of muscular dystrophy predominantly affecting boys. The onset of muscle weakness typically begins around age four, with rapid progression. Initially, muscle loss occurs in the thighs and pe ...
.
**Caused by SVA transposable element insertion in the fukutin (FKTN) gene which renders the gene inactive.
* Alzheimer's Disease and other Tauopathies
** Transposable element dysregulation can cause neuronal death, leading to neurodegenerative disorders
Rate of transposition, induction and defense
One study estimated the rate of transposition of a particular retrotransposon, the Ty1 element in ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
''. Using several assumptions, the rate of successful transposition event per single Ty1 element came out to be about once every few months to once every few years. Some TEs contain heat-shock like promoters and their rate of transposition increases if the cell is subjected to stress, thus increasing the mutation rate under these conditions, which might be beneficial to the cell.
Cells defend against the proliferation of TEs in a number of ways. These include piRNA
Pirna (; , ) is a town in Saxony, Germany and capital of the administrative district Sächsische Schweiz-Osterzgebirge. The town's population is over 37,000. Pirna is located near Dresden and is an important district town as well as a ''Große ...
s and siRNA
Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded non-coding RNA molecules, typically 20–24 base pairs in length, similar to microRNA (miRNA), and operating within the RN ...
s, which silence
Silence is the absence of ambient hearing, audible sound, the emission of sounds of such low sound intensity, intensity that they do not draw attention to themselves, or the state of having ceased to produce sounds; this latter sense can be exten ...
TEs after they have been transcribed.
If organisms are mostly composed of TEs, one might assume that disease caused by misplaced TEs is very common, but in most cases TEs are silenced through epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
mechanisms like DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
, chromatin remodeling and piRNA, such that little to no phenotypic effects nor movements of TEs occur as in some wild-type plant TEs. Certain mutated plants have been found to have defects in methylation-related enzymes (methyl transferase) which cause the transcription of TEs, thus affecting the phenotype.
One hypothesis suggests that only approximately 100 LINE1 related sequences are active, despite their sequences making up 17% of the human genome. In human cells, silencing of LINE1 sequences is triggered by an RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
(RNAi) mechanism. Surprisingly, the RNAi sequences are derived from the 5′ untranslated region (UTR) of the LINE1, a long terminal which repeats itself. Supposedly, the 5′ LINE1 UTR that codes for the sense promoter for LINE1 transcription also encodes the antisense promoter for the miRNA
Micro ribonucleic acid (microRNA, miRNA, μRNA) are small, single-stranded, non-coding RNA molecules containing 21–23 nucleotides. Found in plants, animals, and even some viruses, miRNAs are involved in RNA silencing and post-transcri ...
that becomes the substrate for siRNA production. Inhibition of the RNAi silencing mechanism in this region showed an increase in LINE1 transcription.
Evolution
TEs are found in almost all life forms, and the scientific community is still exploring their evolution and their effect on genome evolution. It is unclear whether TEs originated in thelast universal common ancestor
The last universal common ancestor (LUCA) is the hypothesized common ancestral cell from which the three domains of life, the Bacteria, the Archaea, and the Eukarya originated. The cell had a lipid bilayer; it possessed the genetic code a ...
, arose independently multiple times, or arose once and then spread to other kingdoms by horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the e ...
.
Because excessive TE activity can damage exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence ...
s, many organisms have acquired mechanisms to inhibit their activity. Bacteria may undergo high rates of gene deletion
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleoti ...
as part of a mechanism to remove TEs and viruses from their genomes, while eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
organisms typically use RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
to inhibit TE activity. Nevertheless, some TEs generate large families often associated with speciation
Speciation is the evolutionary process by which populations evolve to become distinct species. The biologist Orator F. Cook coined the term in 1906 for cladogenesis, the splitting of lineages, as opposed to anagenesis, phyletic evolution within ...
events. Evolution often deactivates DNA transposons, leaving them as intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
s (inactive gene sequences). In vertebrate animal cells, nearly all 100,000+ DNA transposons per genome have genes that encode inactive transposase polypeptides. The first synthetic transposon designed for use in vertebrate (including human) cells, the Sleeping Beauty transposon system
The ''Sleeping Beauty'' transposon system is a synthetic DNA transposon designed to introduce precisely defined DNA sequences into the chromosomes of vertebrate animals for the purposes of introducing new traits and to discover new genes and thei ...
, is a Tc1/mariner-like transposon. Its dead ("fossil") versions are spread widely in the salmonid genome and a functional version was engineered by comparing those versions. Human Tc1-like transposons are divided into Hsmar1 and Hsmar2 subfamilies. Although both types are inactive, one copy of Hsmar1 found in the SETMAR gene is under selection as it provides DNA-binding for the histone-modifying protein. Many other human genes are similarly derived from transposons. Hsmar2 has been reconstructed multiple times from the fossil sequences.
The frequency and location of TE integrations influence genomic structure and evolution and affect gene and protein regulatory networks during development and in differentiated cell types. Large quantities of TEs within genomes may still present evolutionary advantages, however. Interspersed repeat Interspersed repetitive DNA is found in all eukaryotic genomes. They differ from tandem repeat DNA in that rather than the repeat sequences coming right after one another, they are dispersed throughout the genome and nonadjacent. The sequence that r ...
s within genomes are created by transposition events accumulating over evolutionary time. Because interspersed repeats block gene conversion
Gene conversion is the process by which one DNA sequence replaces a homologous sequence such that the sequences become identical after the conversion. Gene conversion can be either allelic, meaning that one allele of the same gene replaces another ...
, they protect novel gene sequences from being overwritten by similar gene sequences and thereby facilitate the development of new genes. TEs may also have been co-opted by the vertebrate immune system as a means of producing antibody diversity. The V(D)J recombination
V(D)J recombination (variable–diversity–joining rearrangement) is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire ...
system operates by a mechanism similar to that of some TEs. TEs also serve to generate repeating sequences that can form dsRNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself ( non-coding RNA) or by forming a template for the production of proteins ( messenger RNA). RNA and deoxy ...
to act as a substrate for the action of ADAR
Adar (Hebrew: , ; from Akkadian ''adaru'') is the sixth month of the civil year and the twelfth month of the religious year on the Hebrew calendar, roughly corresponding to the month of March in the Gregorian calendar. It is a month of 29 days. ...
in RNA editing.
TEs can contain many types of genes, including those conferring antibiotic resistance and the ability to transpose to conjugative plasmids. Some TEs also contain integron Integrons are genetic mechanisms that allow bacteria to adapt and evolve rapidly through the stockpiling and expression of new genes. These genes are embedded in a specific genetic structure called gene cassette (a term that is lately changing to i ...
s, genetic elements that can capture and express genes from other sources. These contain integrase
Retroviral integrase (IN) is an enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme ...
, which can integrate gene cassette
In biology, a gene cassette is a type of mobile genetic element that contains a gene and a recombination site. Each cassette usually contains a single gene and tends to be very small; on the order of 500–1,000 base pairs. They may exist incorpor ...
s. There are over 40 antibiotic resistance genes identified on cassettes, as well as virulence genes.
Transposons do not always excise their elements precisely, sometimes removing the adjacent base pairs; this phenomenon is called exon shuffling
Exon shuffling is a molecular mechanism for the formation of new genes. It is a process through which two or more exons from different genes can be brought together ectopically, or the same exon can be duplicated, to create a new exon-intron st ...
. Shuffling two unrelated exons can create a novel gene product or, more likely, an intron.
Some non-autonomous DNA TEs found in plants can capture coding DNA from genes and shuffle them across the genome. This process can duplicate genes in the genome (a phenomenon called transduplication), and can contribute to generate novel genes by exon shuffling.
Evolutionary drive for TEs on the genomic context
There is a hypothesis that states that TEs might provide a ready source of DNA that could be co-opted by the cell to help regulate gene expression. Research showed that many diverse modes of TEs co-evolution along with some transcription factors targeting TE-associated genomic elements and chromatin are evolving from TE sequences. Most of the time, these particular modes do not follow the simple model of TEs and regulating host gene expression.Applications
Transposable elements can be harnessed in laboratory and research settings to study genomes of organisms and even engineer genetic sequences. The use of transposable elements can be split into two categories: for genetic engineering and as a genetic tool.Genetic engineering
* Insertional mutagenesis uses the features of a TE to insert a sequence. In most cases, this is used to remove a DNA sequence or cause a frameshift mutation. ** In some cases the insertion of a TE into a gene can disrupt that gene's function in a reversible manner where transposase-mediated excision of the DNA transposon restores gene function. ** This produces plants in which neighboring cells have differentgenotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a ...
s.
** This feature allows researchers to distinguish between genes that must be present inside of a cell in order to function (cell-autonomous) and genes that produce observable effects in cells other than those where the gene is expressed.
Genetic tool
In addition to the qualities mentioned for Genetic engineering, a Genetic tool also:- * Used for analysis of gene expression and protein functioning in signature-tagging mutagenesis. ** This analytical tool allows researchers the ability to determinephenotypic
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
expression of gene sequences. Also, this analytic technique mutates the desired locus of interest so that the phenotypes of the original and the mutated gene can be compared.
Specific applications
* TEs are also a widely used tool for mutagenesis of most experimentally tractable organisms. The Sleeping Beauty transposon system has been used extensively as an insertional tag for identifying cancer genes. * The Tc1/mariner-class of TEs Sleeping Beauty transposon system, awarded Molecule of the Year in 2009, is active in mammalian cells and is being investigated for use in human gene therapy. * TEs are used for the reconstruction of phylogenies by the means of presence/absence analyses. Transposons can act as biological mutagen in bacteria. * Common organisms which the use of Transposons has been well developed are: **''Drosophila
''Drosophila'' (), from Ancient Greek δρόσος (''drósos''), meaning "dew", and φίλος (''phílos''), meaning "loving", is a genus of fly, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or p ...
''
** ''Arabidopsis thaliana
''Arabidopsis thaliana'', the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally ...
''
** ''Escherichia coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus '' Escherichia'' that is commonly fo ...
''
''De novo'' repeat identification
''De novo'' repeat identification is an initial scan of sequence data that seeks to find the repetitive regions of the genome, and to classify these repeats. Many computer programs exist to perform ''de novo'' repeat identification, all operating under the same general principles. As short tandem repeats are generally 1–6 base pairs in length and are often consecutive, their identification is relatively simple. Dispersed repetitive elements, on the other hand, are more challenging to identify, due to the fact that they are longer and have often acquired mutations. However, it is important to identify these repeats as they are often found to be transposable elements (TEs). ''De novo'' identification of transposons involves three steps: 1) find all repeats within the genome, 2) build a consensus of each family of sequences, and 3) classify these repeats. There are three groups of algorithms for the first step. One group is referred to as thek-mer
In bioinformatics, ''k''-mers are substrings of length k contained within a biological sequence. Primarily used within the context of computational genomics and sequence analysis, in which ''k''-mers are composed of nucleotides (''i.e''. A, T, ...
approach, where a k-mer is a sequence of length k. In this approach, the genome is scanned for overrepresented k-mers; that is, k-mers that occur more often than is likely based on probability alone. The length k is determined by the type of transposon being searched for. The k-mer approach also allows mismatches, the number of which is determined by the analyst. Some k-mer approach programs use the k-mer as a base, and extend both ends of each repeated k-mer until there is no more similarity between them, indicating the ends of the repeats. Another group of algorithms employs a method called sequence self-comparison. Sequence self-comparison programs use databases such as AB-BLAST to conduct an initial sequence alignment
In bioinformatics, a sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural biology, structural, or evolutionary relationships between ...
. As these programs find groups of elements that partially overlap, they are useful for finding highly diverged transposons, or transposons with only a small region copied into other parts of the genome. Another group of algorithms follows the periodicity approach. These algorithms perform a Fourier transformation
In mathematics, the Fourier transform (FT) is an integral transform that takes a function as input then outputs another function that describes the extent to which various frequencies are present in the original function. The output of the tr ...
on the sequence data, identifying periodicities, regions that are repeated periodically, and are able to use peaks in the resultant spectrum to find candidate repetitive elements. This method works best for tandem repeats, but can be used for dispersed repeats as well. However, it is a slow process, making it an unlikely choice for genome-scale analysis.
The second step of ''de novo'' repeat identification involves building a consensus of each family of sequences. A consensus sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated sequence of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It represents the result ...
is a sequence that is created based on the repeats that comprise a TE family. A base pair in a consensus is the one that occurred most often in the sequences being compared to make the consensus. For example, in a family of 50 repeats where 42 have a T base pair in the same position, the consensus sequence would have a T at this position as well, as the base pair is representative of the family as a whole at that particular position, and is most likely the base pair found in the family's ancestor at that position. Once a consensus sequence has been made for each family, it is then possible to move on to further analysis, such as TE classification and genome masking in order to quantify the overall TE content of the genome.
Adaptive TEs
Transposable elements have been recognized as good candidates for stimulating gene adaptation, through their ability to regulate the expression levels of nearby genes. Combined with their "mobility", transposable elements can be relocated adjacent to their targeted genes, and control the expression levels of the gene, dependent upon the circumstances. The study conducted in 2008, "High Rate of Recent Transposable Element–Induced Adaptation in Drosophila melanogaster", used ''D. melanogaster'' that had recently migrated from Africa to other parts of the world, as a basis for studying adaptations caused by transposable elements. Although most of the TEs were located on introns, the experiment showed a significant difference in gene expressions between the population in Africa and other parts of the world. The four TEs that caused the selective sweep were more prevalent in ''D. melanogaster'' from temperate climates, leading the researchers to conclude that the selective pressures of the climate prompted genetic adaptation. From this experiment, it has been confirmed that adaptive TEs are prevalent in nature, by enabling organisms to adapt gene expression as a result of new selective pressures. However, not all effects of adaptive TEs are beneficial to the population. In the research conducted in 2009, "A Recent Adaptive Transposable Element Insertion Near Highly Conserved Developmental Loci in Drosophila melanogaster", a TE, inserted between Jheh 2 and Jheh 3, revealed a downgrade in the expression level of both of the genes. Downregulation of such genes has caused ''Drosophila'' to exhibit extended developmental time and reduced egg to adult viability. Although this adaptation was observed in high frequency in all non-African populations, it was not fixed in any of them. This is not hard to believe, since it is logical for a population to favor higher egg to adult viability, therefore trying to purge the trait caused by this specific TE adaptation. At the same time, there have been several reports showing the advantageous adaptation caused by TEs. In the research done with silkworms, "An Adaptive Transposable Element insertion in the Regulatory Region of the EO Gene in the Domesticated Silkworm", a TE insertion was observed in the cis-regulatory region of the EO gene, which regulates molting hormone 20E, and enhanced expression was recorded. While populations without the TE insert are often unable to effectively regulate hormone 20E under starvation conditions, those with the insert had a more stable development, which resulted in higher developmental uniformity. These three experiments all demonstrated different ways in which TE insertions can be advantageous or disadvantageous, through means of regulating the expression level of adjacent genes. The field of adaptive TE research is still under development and more findings can be expected in the future.TEs participates in gene control networks
Recent studies have confirmed that TEs can contribute to the generation of transcription factors. However, how this process of contribution can have an impact on the participation of genome control networks. TEs are more common in many regions of the DNA and it makes up 45% of total human DNA. Also, TEs contributed to 16% of transcription factor binding sites. A larger number of motifs are also found in non-TE-derived DNA, and the number is larger than TE-derived DNA. All these factors correlate to the direct participation of TEs in many ways of gene control networks.See also
Notes
* * *References
External links
* – A possible connection between aberrant reinsertions and lymphoma.Repbase
– a database of transposable element sequences
Dfam
- a database of transposable element families, multiple sequence alignments, and sequence models
RepeatMasker
– a computer program used by computational biologists to
annotate
An annotation is extra information associated with a particular point in a document or other piece of information. It can be a note that includes a comment or explanation. Annotations are sometimes presented in the margin of book pages. For anno ...
transposons in DNA sequences
Use of the Sleeping Beauty Transposon System for Stable Gene Expression in Mouse Embryonic Stem Cells
Introduction to Transposons, 2018 YouTube video
{{Authority control Modification of genetic information Mobile genetic elements Molecular biology Non-coding DNA