Transition Metal Imidazole Complex on:
[Wikipedia]
[Google]
[Amazon]
 A transition metal imidazole complex is a
A transition metal imidazole complex is a
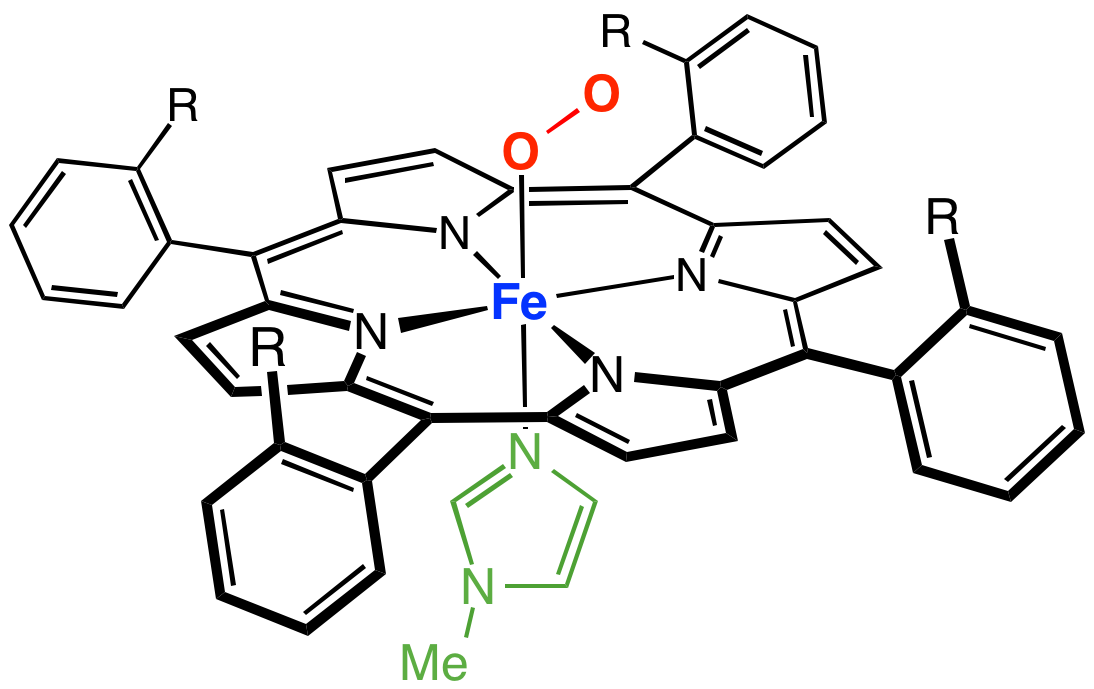 Only the
Only the
 A transition metal imidazole complex is a
A transition metal imidazole complex is a coordination complex
A coordination complex is a chemical compound consisting of a central atom or ion, which is usually metallic and is called the ''coordination centre'', and a surrounding array of chemical bond, bound molecules or ions, that are in turn known as ' ...
that has one or more imidazole
Imidazole (ImH) is an organic compound with the formula . It is a white or colourless solid that is soluble in water, producing a mildly alkaline solution. It can be classified as a heterocycle, specifically as a diazole.
Many natural products, ...
ligand
In coordination chemistry, a ligand is an ion or molecule with a functional group that binds to a central metal atom to form a coordination complex. The bonding with the metal generally involves formal donation of one or more of the ligand's el ...
s. Complexes of imidazole itself are of little practical importance. In contrast, imidazole derivatives, especially histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an Amine, α-amino group (which is in the protonated –NH3+ form under Physiological condition, biological conditions), a carboxylic ...
, are pervasive ligands in biology where they bind metal cofactors.
Bonding and structure
: Only the
Only the imine
In organic chemistry, an imine ( or ) is a functional group or organic compound containing a carbon–nitrogen double bond (). The nitrogen atom can be attached to a hydrogen or an organic group (R). The carbon atom has two additional single bon ...
nitrogen (HC=N-CH) of imidazole is basic, and it is this nitrogen that binds to metal ions. Imidazole is a pure sigma-donor ligand. The pKa of protonated imidazolium
Imidazole (ImH) is an organic compound with the formula . It is a white or colourless solid that is soluble in water, producing a mildly alkaline solution. It can be classified as a heterocycle, specifically as a diazole.
Many natural products, ...
cation is about 6.95, which indicates that the basicity of imidazole is intermediate between pyridine
Pyridine is a basic (chemistry), basic heterocyclic compound, heterocyclic organic compound with the chemical formula . It is structurally related to benzene, with one methine group replaced by a nitrogen atom . It is a highly flammable, weak ...
(pKa of pyridinium = 5.23) and ammonia (pKa = 9,24 of ammonium). The donor properties of imidazole can also inferred from the redox properties of its complexes. It is classified as an L ligand in the Covalent bond classification method The covalent bond classification (CBC) method, also referred to as LXZ notation, is a way of describing covalent compounds such as organometallic complexes in a way that is not prone to limitations resulting from the definition of oxidation state. ...
. In the usual electron counting method, it is a two-electron ligand.
Imidazole can be classified as hard
Hard means something that is difficult to do. It may also refer to:
* Hardness, resistance of physical materials to deformation or fracture
* Hard water, water with high mineral content
Arts and entertainment
* Hard (TV series), ''Hard'' (TV ser ...
ligand. Nonetheless, complexes between low-valent metals and imidazole are well known, e.g., e(imidazole)3(CO)3sup>+.
Imidazole is a compact, flat ligand. Six imidazole ligands fit comfortably around octahedral metal centers, e.g., e(imidazole)6sup>2+. The M-N(imidazole) bond is freely rotating.
Homoleptic octahedral complexes have been characterized by X-ray crystallography for the following dications: Fe2+, Co2+, Ni2+, Zn2+, Cd2+. Hexakis complexes of both Ru2+ and Ru3+ are also known. Cu2+, Pd2+, and Pt2+ form homoleptic square planar complexes. Zn2+, although crystallized as the hexakis complex, more typically forms a tetrahedral complex.
Complexes of substituted imidazoles
Structure of vitamin b12, illustrating the dimethylbenzimidazole ligand N-methylimidazole is slightly more basic than imidazole but is otherwise similar, if more lipophilic. Many salts of (imidazole-1-R)6sup>2+ are known (R = alkyl, vinyl, etc.).2-Methylimidazole
2-Methylimidazole is an organic compound that is structurally related to imidazole with the chemical formula CH3C3H2N2H. It is a white or colorless solid that is highly soluble in polar organic solvents and water. It is a precursor to a range of d ...
s are somewhat bulky ligands owing to the steric clash between the 2-methyl group and other ligands in octahedral complexes.
A modified benzimidazole
Benzimidazole is a heterocyclic aromatic organic compound. This bicyclic compound may be viewed as fused rings of the aromatic compounds benzene and imidazole. It is a white solid that appears in form of tabular crystals.
Preparation
Benzimi ...
ligand is found in all versions of vitamin B12
Vitamin B12, also known as cobalamin, is a water-soluble vitamin involved in metabolism. One of eight B vitamins, it serves as a vital cofactor (biochemistry), cofactor in DNA synthesis and both fatty acid metabolism, fatty acid and amino a ...
.
Histidine
Histidine complexes comprise an important subset of transition metal amino acid complexes. In common with other 3-substituted imidazoles, histidine can coordinate to metals via either of two nonequivalenttautomers
In chemistry, tautomers () are structural isomers (constitutional isomers) of chemical compounds that readily interconvert.
The chemical reaction interconverting the two is called tautomerization. This conversion commonly results from the reloca ...
. The free amino acid can coordinate through the imidazole and either or both of the carboxylate and amine.
The imidazole side chain of histidine residues in protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s are common binding sites for metal ions. Unlike the free amino acid, the histidine residue (i.e., as a component of a peptide or protein), coordinates solely via the imidazole substituent. Examples include myoglobin
Myoglobin (symbol Mb or MB) is an iron- and oxygen-binding protein found in the cardiac and skeletal muscle, skeletal Muscle, muscle tissue of vertebrates in general and in almost all mammals. Myoglobin is distantly related to hemoglobin. Compar ...
(Fe), carbonic anhydrase
The carbonic anhydrases (or carbonate dehydratases) () form a family of enzymes that catalyst, catalyze the interconversion between carbon dioxide and water and the Dissociation (chemistry), dissociated ions of carbonic acid (i.e. bicarbonate a ...
(Zn), azurin
Azurin is a small, periplasmic, bacterial blue copper protein found in ''Pseudomonas'', ''Bordetella'', or ''Alcaligenes'' bacteria. Azurin moderates single-electron transfer between enzymes associated with the cytochrome chain by undergoing oxida ...
(Cu), and alpha-ketoglutarate-dependent hydroxylases Alpha-ketoglutarate-dependent hydroxylases are a major class of non-heme iron proteins that catalyse a wide range of reactions. These reactions include hydroxylation reactions, demethylations, ring expansions, ring closures, and desaturations. Func ...
(Fe). Polyhistidine-tag
A polyhistidine-tag, best known by the trademarked name His-tag, is an amino acid motif in proteins that typically consists of at least six histidine (''His'') residues, often at the N- or C-terminus of the protein. It is also known as a hexa his ...
("his tag") is an amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
motif in protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s consisting of several histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an Amine, α-amino group (which is in the protonated –NH3+ form under Physiological condition, biological conditions), a carboxylic ...
(''His'') residues that is attached to proteins to facilitate purification. The concept relies on the affinity of the imidazole side chain for metal cations.
Reactions of imidazole ligands
Especially in cationic imidazole complexes, the N-H center is acidified. For tricationic d6 pentammines, deprotonation of the imidazole ligand gives imidazolate complexes with pKa near 10 (M = Co, Rh, Ir): : (NH3)5(N2C3H4)sup>3+ (NH3)5(N2C3H3)sup>2+ + H+ The d5 complex u(NH3)5(N2C3H4)sup>3+ is more acidic, with a pKa of 8.9. Thus, complexation to tricationic complexes acidify the pyrrolic NH center by at least 10,000. Imidazole ligands are isomers ofN-heterocyclic carbene
A persistent carbene (also known as stable carbene) is an organic molecule whose natural resonance structure has a carbon atom with octet rule, incomplete octet (a carbene), but does not exhibit the tremendous instability typically associated with ...
s. This conversion has been observed:
: u(NH3)5(N2C3H4)sup>2+ → u(NH3)5(C(NH)2(CH)2)sup>2+
Imidazolate complexes
The pKa of imidazole (to give imidazolate) is 14, thus it is easier to deprotonate than many otheramine
In chemistry, amines (, ) are organic compounds that contain carbon-nitrogen bonds. Amines are formed when one or more hydrogen atoms in ammonia are replaced by alkyl or aryl groups. The nitrogen atom in an amine possesses a lone pair of elec ...
s or imine
In organic chemistry, an imine ( or ) is a functional group or organic compound containing a carbon–nitrogen double bond (). The nitrogen atom can be attached to a hydrogen or an organic group (R). The carbon atom has two additional single bon ...
s. Many metal complexes feature imidazolate as a bridging ligand
In coordination chemistry, a bridging ligand is a ligand that connects two or more atoms, usually metal ions. The ligand may be atomic or polyatomic. Virtually all complex organic compounds can serve as bridging ligands, so the term is usually r ...
. One example of an imidazolate complex from biochemistry is found at the active site of copper-containing superoxide dismutase
Superoxide dismutase (SOD, ) is an enzyme that alternately catalyzes the dismutation (or partitioning) of the superoxide () anion radical into normal molecular oxygen (O2) and hydrogen peroxide (). Superoxide is produced as a by-product of oxy ...
.
The M2(μ-imidazolate) motif underpins materials comprising zeolitic imidazolate framework
Zeolitic imidazolate frameworks (ZIFs) are a class of metal-organic frameworks (MOFs) that are topologically isomorphic with zeolites. ZIFs are composed of tetrahedrally-coordinated transition metal ions (e.g. Fe, Co, Zn) connected by imida ...
s ("ZIF"s).
References