Quaternary Structure on:
[Wikipedia]
[Google]
[Amazon]
Protein quaternary structure is the fourth (and highest) classification level of
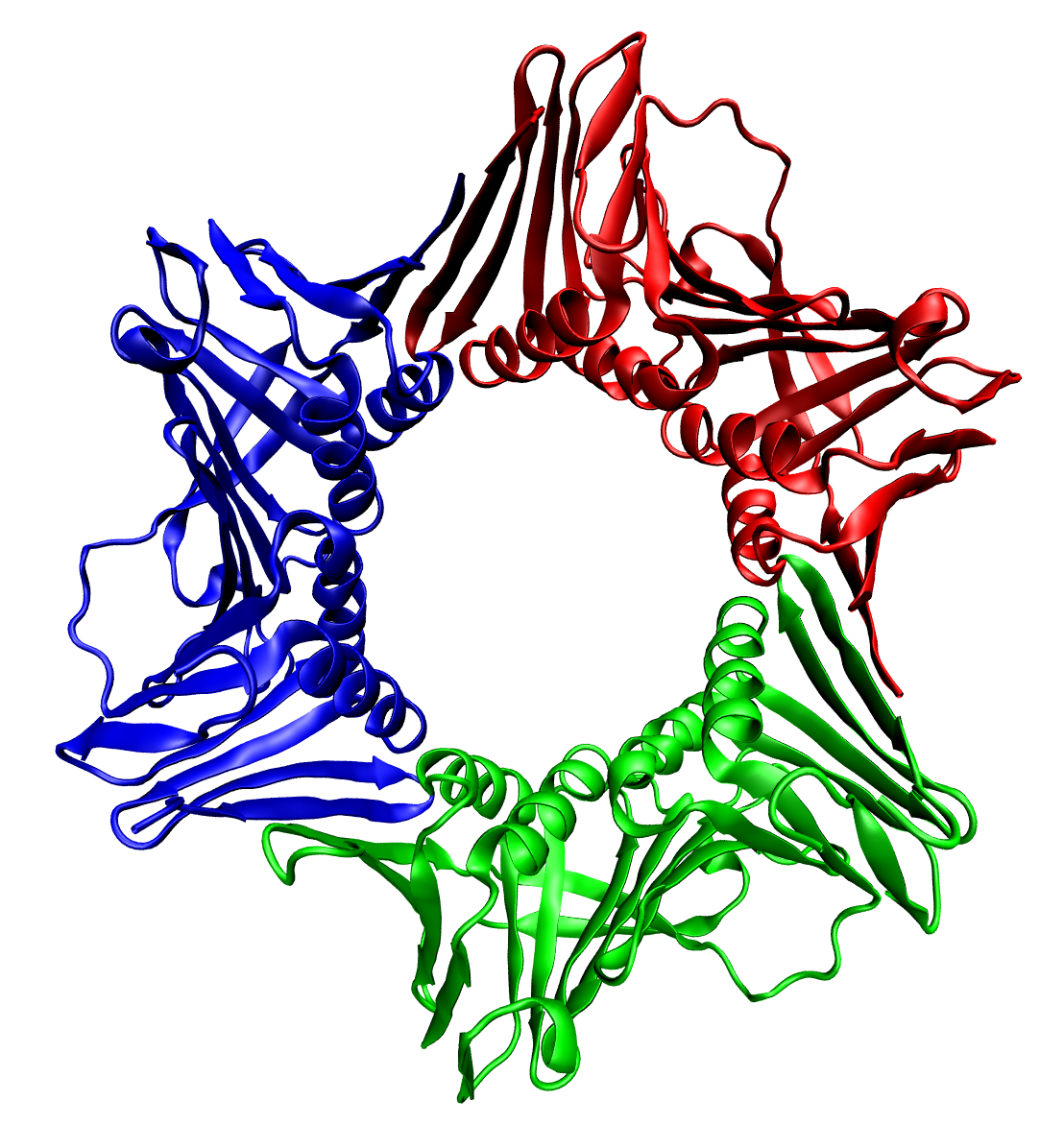 The number of subunits in an oligomeric complex is described using names that end in -mer (Greek for "part, subunit"). Formal and Greco-Latinate names are generally used for the first ten types and can be used for up to twenty subunits, whereas higher order complexes are usually described by the number of subunits, followed by -meric.
:
The number of subunits in an oligomeric complex is described using names that end in -mer (Greek for "part, subunit"). Formal and Greco-Latinate names are generally used for the first ten types and can be used for up to twenty subunits, whereas higher order complexes are usually described by the number of subunits, followed by -meric.
:* ''No known examples''
The smallest unit forming a homo-oligomer, i.e. one protein chain or subunit, is designated as a monomer, subunit or protomer. The latter term was originally devised to specify the smallest unit of hetero-oligomeric proteins, but is also applied to homo-oligomeric proteins in current literature. The subunits usually arrange in cyclic symmetry to form closed
The Macromolecular Structure Database
(MSD) at the European Bioinformatics Institute (EBI) – Serves a list of the Probable Quaternary Structure (PQS) for every protein in the
PQS server
– PQS has not been updated since August 2009
– The Protein Interfaces, Surfaces and Assemblies server at the MSD.
EPPIC
– Evolutionary Protein–Protein Interface Classification: evolutionary assessment of interfaces in crystal structures
3D complex
– Structural classification of protein complexes * Proteopedia â€
Proteopedia Home Page
The collaborative, 3D encyclopedia of proteins and other molecules. * PDBWiki â€
PDBWiki Home Page
– a website for community annotation of PDB structures. * ProtCID â€
ProtCID
€”a database of similar protein–protein interfaces in crystal structures of homologous proteins. {{Biomolecular structure Protein structure 4 Stereochemistry
protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid ...
. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also referred to as subunits). Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acid
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a pentose, 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nuclei ...
s and other cofactors.
Description and examples
Many proteins are actually assemblies of multiplepolypeptide
Peptides are short chains of amino acids linked by peptide bonds. A polypeptide is a longer, continuous, unbranched peptide chain. Polypeptides that have a molecular mass of 10,000 Da or more are called proteins. Chains of fewer than twenty ...
chains. The quaternary structure refers to the number and arrangement of the protein subunits with respect to one another. Examples of proteins with quaternary structure include hemoglobin
Hemoglobin (haemoglobin, Hb or Hgb) is a protein containing iron that facilitates the transportation of oxygen in red blood cells. Almost all vertebrates contain hemoglobin, with the sole exception of the fish family Channichthyidae. Hemoglobin ...
, DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
, ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
s, antibodies
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as bacteria and viruses, including those that caus ...
, and ion channel
Ion channels are pore-forming membrane proteins that allow ions to pass through the channel pore. Their functions include establishing a resting membrane potential, shaping action potentials and other electrical signals by Gating (electrophysiol ...
s.
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s composed of subunits with diverse functions are sometimes called holoenzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s, in which some parts may be known as regulatory subunits and the functional core is known as the catalytic subunit. Other assemblies referred to instead as multiprotein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain.
Protein c ...
es also possess quaternary structure. Examples include nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
s and microtubule
Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can be as long as 50 micrometres, as wide as 23 to 27 nanometer, nm and have an inner diameter bet ...
s. Changes in quaternary structure can occur through conformational changes within individual subunits or through reorientation of the subunits relative to each other. It is through such changes, which underlie cooperativity and allostery in "multimeric" enzymes, that many proteins undergo regulation and perform their physiological function.
The above definition follows a classical approach to biochemistry, established at times when the distinction between a protein and a functional, proteinaceous unit was difficult to elucidate. More recently, people refer to protein–protein interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and t ...
when discussing quaternary structure of proteins and consider all assemblies of proteins as protein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple active site, catalytic domains are found in a single polypeptide chain.
...
es.
Nomenclature
 The number of subunits in an oligomeric complex is described using names that end in -mer (Greek for "part, subunit"). Formal and Greco-Latinate names are generally used for the first ten types and can be used for up to twenty subunits, whereas higher order complexes are usually described by the number of subunits, followed by -meric.
:
The number of subunits in an oligomeric complex is described using names that end in -mer (Greek for "part, subunit"). Formal and Greco-Latinate names are generally used for the first ten types and can be used for up to twenty subunits, whereas higher order complexes are usually described by the number of subunits, followed by -meric.
:point group
In geometry, a point group is a group (mathematics), mathematical group of symmetry operations (isometry, isometries in a Euclidean space) that have a Fixed point (mathematics), fixed point in common. The Origin (mathematics), coordinate origin o ...
symmetries.
Although complexes higher than octamers are rarely observed for most proteins, there are some important exceptions. Viral capsids are often composed of multiples of 60 proteins. Several molecular machines are also found in the cell, such as the proteasome (four heptameric rings = 28 subunits), the transcription complex and the spliceosome. The ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
is probably the largest molecular machine, and is composed of many RNA and protein molecules.
In some cases, proteins form complexes that then assemble into even larger complexes. In such cases, one uses the nomenclature, e.g., "dimer of dimers" or "trimer of dimers". This may suggest that the complex might dissociate into smaller sub-complexes before dissociating into monomers. This usually implies that the complex consists of different oligomerisation interfaces. For example, a tetrameric protein may have one four-fold rotation axis, i.e. point group symmetry 4 or ''C''4. In this case the four interfaces between the subunits are identical. It may also have point group symmetry 222 or ''D''2. This tetramer has different interfaces and the tetramer can dissociate into two identical homodimers. Tetramers of 222 symmetry are "dimer of dimers". Hexamers of 32 point group symmetry are "trimer of dimers" or "dimer of trimers". Thus, the nomenclature "dimer of dimers" is used to specify the point group symmetry or arrangement of the oligomer, independent of information relating to its dissociation properties.
Another distinction often made when referring to oligomers is whether they are homomeric or heteromeric, referring to whether the smaller protein subunits that come together to make the protein complex are the same (homomeric) or different (heteromeric) from each other. For example, two identical protein monomers would come together to form a homo-dimer, whereas two different protein monomers would create a hetero-dimer.
Structure Determination
Protein quaternary structure can be determined using a variety of experimental techniques that require a sample of protein in a variety of experimental conditions. The experiments often provide an estimate of the mass of the native protein and, together with knowledge of the masses and/or stoichiometry of the subunits, allow the quaternary structure to be predicted with a given accuracy. It is not always possible to obtain a precise determination of the subunit composition for a variety of reasons. The number of subunits in a protein complex can often be determined by measuring the hydrodynamic molecular volume or mass of the intact complex, which requires native solution conditions. For ''folded'' proteins, the mass can be inferred from its volume using the partial specific volume of 0.73 ml/g. However, volume measurements are less certain than mass measurements, since ''unfolded'' proteins appear to have a much larger volume than folded proteins; additional experiments are required to determine whether a protein is unfolded or has formed an oligomer.Common techniques used to study protein quaternary structure
* Ultracentrifugation * Surface-induced dissociation mass spectrometry * Coimmunoprecipation * FRET * Nuclear Magnetic Resonance (NMR)Direct mass measurement of intact complexes
* Sedimentation-equilibrium analytical ultracentrifugation * Electrospraymass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used ...
* Mass Spectrometric Immunoassay MSIA
Direct size measurement of intact complexes
* Static light scattering * Size exclusion chromatography (requires calibration) * Dual polarisation interferometryIndirect size measurement of intact complexes
* Sedimentation-velocity analytical ultracentrifugation (measures the translational diffusion constant) * Dynamic light scattering (measures the translational diffusion constant) * Pulsed-gradient protein nuclear magnetic resonance (measures the translational diffusion constant) * Fluorescence polarization (measures the rotational diffusion constant) * Dielectric relaxation (measures the rotational diffusion constant) * Dual polarisation interferometry (measures the size and the density of the complex) Methods that measure the mass or volume under unfolding conditions (such as MALDI-TOFmass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a ''mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is used ...
and SDS-PAGE) are generally not useful, since non-native conditions usually cause the complex to dissociate into monomers. However, these may sometimes be applicable; for example, the experimenter may apply SDS-PAGE after first treating the intact complex with chemical cross-link
In chemistry and biology, a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural ...
reagents.
Structure Prediction
Some bioinformatics methods have been developed for predicting the quaternary structural attributes of proteins based on their sequence information by using various modes of pseudo amino acid composition. Protein folding prediction programs used to predict protein tertiary structure have also been expanding to better predict protein quaternary structure. One such development is AlphaFold-Multimer built upon the AlphaFold model for predicting protein tertiary structure.Role in Cell Signaling
Protein quaternary structure also plays an important role in certain cell signaling pathways. The G-protein coupled receptor pathway involves a heterotrimeric protein known as a G-protein. G-proteins contain three distinct subunits known as the G-alpha, G-beta, and G-gamma subunits. When the G-protein is activated, it binds to the G-protein coupled receptor protein and the cell signaling pathway is initiated. Another example is the receptor tyrosine kinase (RTK) pathway, which is initiated by the dimerization of two receptor tyrosine kinase monomers. When the dimer is formed, the two kinases can phosphorylate each other and initiate a cell signaling pathway.Protein–protein interactions
Proteins are capable of forming very tight but also only transient complexes. For example, ribonuclease inhibitor binds to ribonuclease A with a roughly 20 fM dissociation constant. Other proteins have evolved to bind specifically to unusual moieties on another protein, e.g., biotin groups (avidin), phosphorylated tyrosines ( SH2 domains) or proline-rich segments (SH3 domain
The SRC Homology 3 Domain (or SH3 domain) is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of ph ...
s). Protein–protein interactions can be engineered to favor certain oligomerization states.
Intragenic complementation
When multiple copies of a polypeptide encoded by agene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
form a quaternary complex, this protein structure is referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant
In biology, and especially in genetics, a mutant is an organism or a new genetic character arising or resulting from an instance of mutation, which is generally an alteration of the DNA sequence of the genome or chromosome of an organism. It i ...
allele
An allele is a variant of the sequence of nucleotides at a particular location, or Locus (genetics), locus, on a DNA molecule.
Alleles can differ at a single position through Single-nucleotide polymorphism, single nucleotide polymorphisms (SNP), ...
s of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. In such a case, the phenomenon is referred to as intragenic complementation (also called inter-allelic complementation). Intragenic complementation appears to be common and has been studied in many different genes in a variety of organisms including the fungi '' Neurospora crassa'', ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
'' and '' Schizosaccharomyces pombe''; the bacterium ''Salmonella
''Salmonella'' is a genus of bacillus (shape), rod-shaped, (bacillus) Gram-negative bacteria of the family Enterobacteriaceae. The two known species of ''Salmonella'' are ''Salmonella enterica'' and ''Salmonella bongori''. ''S. enterica'' ...
typhimurium''; the virus bacteriophage T4, an RNA virus, and humans. The intermolecular forces likely responsible for self-recognition and multimer formation were discussed by Jehle.
Assembly
Direct interaction of two nascent proteins emerging from nearbyribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
s appears to be a general mechanism for oligomer formation. Hundreds of protein oligomers were identified that assemble in human cells by such an interaction. The most prevalent form of interaction was between the N-terminal regions of the interacting proteins. Dimer formation appears to be able to occur independently of dedicated assembly machines.
See also
*Structural biology
Structural biology deals with structural analysis of living material (formed, composed of, and/or maintained and refined by living cells) at every level of organization.
Early structural biologists throughout the 19th and early 20th centuries we ...
* Nucleic acid quaternary structure
* Multiprotein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain.
Protein c ...
* Biomolecular complex
* Oligomers
Notes
References
External links
The Macromolecular Structure Database
(MSD) at the European Bioinformatics Institute (EBI) – Serves a list of the Probable Quaternary Structure (PQS) for every protein in the
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained a ...
(PDB).
PQS server
– PQS has not been updated since August 2009
– The Protein Interfaces, Surfaces and Assemblies server at the MSD.
EPPIC
– Evolutionary Protein–Protein Interface Classification: evolutionary assessment of interfaces in crystal structures
3D complex
– Structural classification of protein complexes * Proteopedia â€
Proteopedia Home Page
The collaborative, 3D encyclopedia of proteins and other molecules. * PDBWiki â€
PDBWiki Home Page
– a website for community annotation of PDB structures. * ProtCID â€
ProtCID
€”a database of similar protein–protein interfaces in crystal structures of homologous proteins. {{Biomolecular structure Protein structure 4 Stereochemistry