Primer Extension on:
[Wikipedia]
[Google]
[Amazon]
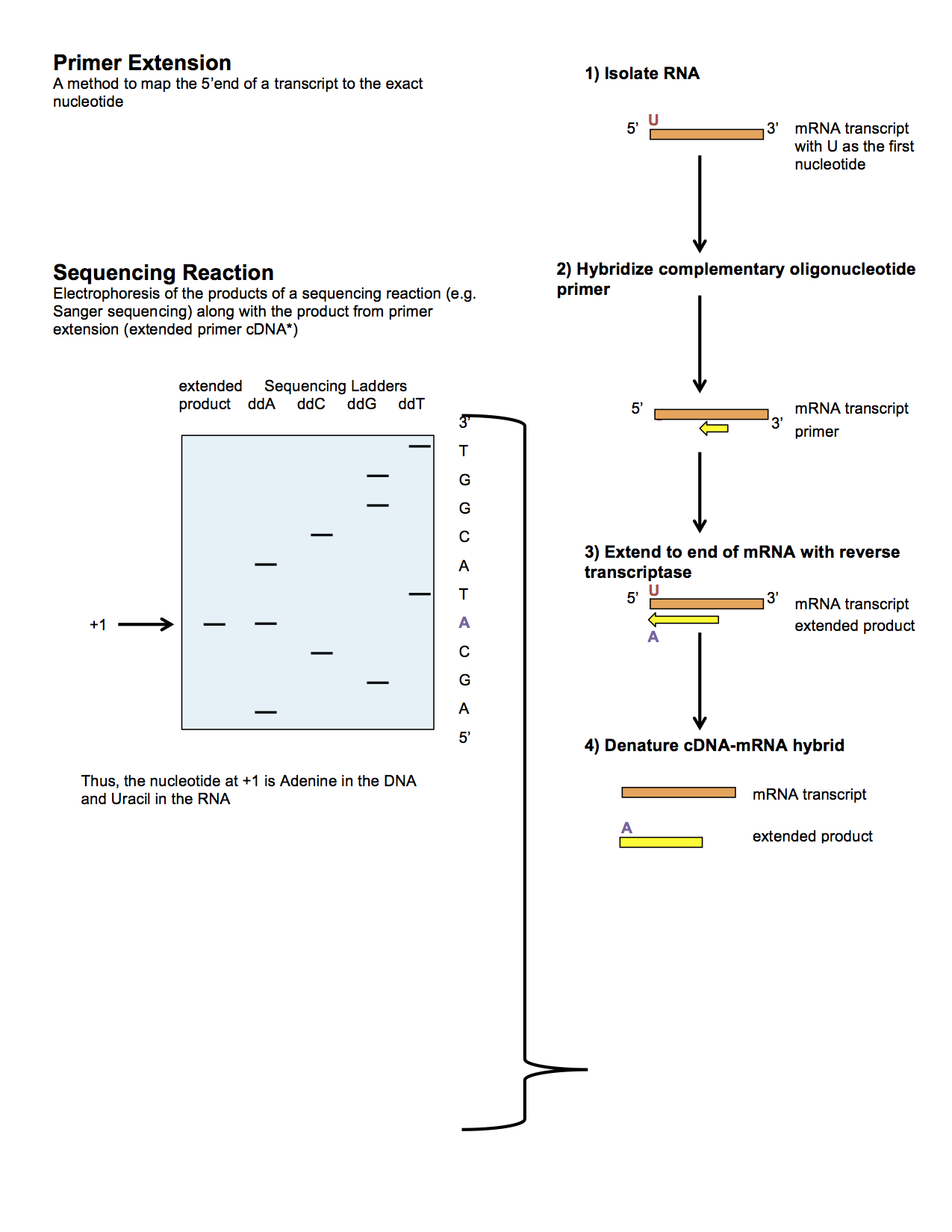 Primer extension is a technique whereby the
Primer extension is a technique whereby the
 Primer extension is a technique whereby the
Primer extension is a technique whereby the 5' end
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-r ...
s of RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
can be mapped - that is, they can be sequenced and properly identified.
Primer extension can be used to determine the start site of transcription (the end site cannot be determined by this method) by which its sequence is known. This technique requires a radiolabelled primer (usually 20 - 50 nucleotides in length) which is complementary to a region near the 3' end of the mRNA. The primer is allowed to anneal to the RNA and reverse transcriptase is used to synthesize cDNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engin ...
from the RNA until it reaches the 5' end of the RNA. By denaturing the hybrid and using the extended primer cDNA as a marker on an electrophoretic gel, it is possible to determine the transcriptional start site. It is usually done so by comparing its location on the gel with the DNA sequence (e.g. Sanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
), preferably by using the same primer on the DNA template strand. The exact nucleotide by which the transcription starts at can be pinpointed by matching the labelled extended primer with the marker nucleotide, who are both sharing the same migration distance on the gel.
Primer extension offers an alternative to a nuclease protection assay
Nuclease protection assay is a laboratory technique used in biochemistry and genetics to identify individual RNA molecules in a heterogeneous RNA sample extracted from cell (biology), cells. The technique can identify one or more RNA molecules of ...
(S1 nuclease mapping) for quantifying and mapping RNA transcripts. The hybridization probe
In molecular biology, a hybridization probe (HP) is a fragment of DNA or RNA, usually 15–10000 nucleotides long, which can be radioactively or fluorescently labeled. HPs can be used to detect the presence of nucleotide sequences in analyzed ...
for primer extension is a synthesized oligonucleotide, whereas S1 mapping requires isolation of a DNA fragment. Both methods provide information where a mRNA starts and provide an estimate of the concentration of a transcript by the intensity of the transcript band on the resulting autoradiograph. Unlike S1 mapping, however, primer extension can only be used to locate the 5’-end of an mRNA transcript because the DNA synthesis required for the assay relies on reverse transcriptase (only polymerizes in the 5’ → 3’ direction).
Primer extension is unaffected by splice sites and is thus preferable in situations where intervening splice sites prevent S1 mapping. Finally, primer extension is more accurate than S1 mapping because the S1 nuclease
Nuclease S1 () is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalysis, catalyses the following chemical reaction
: Endonucleolytic cleavage to 5'-phosphomononucleotide and ...
used in S1 mapping can “nibble off” ends of the RNA-DNA hybrid or fail to degrade the single-stranded regions completely, making a transcript either appear shorter or longer.
References
*https://www.nationaldiagnostics.com/electrophoresis/article/primer-extension *Shenk, T. E., C. Rhodes, P. W. J. Rigby, and P. Berg. “Biochemical Procedure for Production of Small Deletions in Simian Virus 40 DNA.” Proceedings of the National Academy of Sciences 72.4 (1975): 1392-396. Print. Molecular genetics {{Molecular-cell-biology-stub