Niche (protein Structural Motif) on:
[Wikipedia]
[Google]
[Amazon]
In the area of 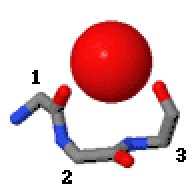 Niches are of two kinds, distinguished as niche3 (3 residues, ''i'' to ''i+2'') and niche4 (4 residues, ''i'' to ''i+3''). In a niche3 motif the δ+-binding carbonyl groups are from residues ''i'' and ''i+2'' while in a niche4 motif they are from residues ''i'' and ''i+3''.
A niche3 has the α conformation for residue ''i+1'' and the β conformation for residue ''i+2''; a niche4 has the α conformation for residues ''i+1'' and ''i+2'' and the β conformation for residue ''i+3''.
A niche occurs commonly at the
Niches are of two kinds, distinguished as niche3 (3 residues, ''i'' to ''i+2'') and niche4 (4 residues, ''i'' to ''i+3''). In a niche3 motif the δ+-binding carbonyl groups are from residues ''i'' and ''i+2'' while in a niche4 motif they are from residues ''i'' and ''i+3''.
A niche3 has the α conformation for residue ''i+1'' and the β conformation for residue ''i+2''; a niche4 has the α conformation for residues ''i+1'' and ''i+2'' and the β conformation for residue ''i+3''.
A niche occurs commonly at the
* PDBeMotif
{{cite journal , last=Golovin , first=A , author2=Henrick , year=2008 , title=MSDmotif: exploring protein sites and motifs , journal=BMC Bioinformatics , volume=9 , issue=1 , pages=312 , doi=10.1186/1471-2105-9-312 , pmc=2491636 , pmid=18637174 , doi-access=free Protein structural motifs
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
structural motifs, niches are three or four amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
residue features in which main-chain CO groups are bridged by positively charged or δ+ groups. The δ+ groups include groups with two hydrogen bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
donor atoms such as NH2 groups and water molecules. In typical proteins, 7% of amino acid residues belong to niches bound to a δ+ group, while another 7% have the conformation but no single cationic
An ion () is an atom or molecule with a net electrical charge. The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by convent ...
bridging group is detected.
 Niches are of two kinds, distinguished as niche3 (3 residues, ''i'' to ''i+2'') and niche4 (4 residues, ''i'' to ''i+3''). In a niche3 motif the δ+-binding carbonyl groups are from residues ''i'' and ''i+2'' while in a niche4 motif they are from residues ''i'' and ''i+3''.
A niche3 has the α conformation for residue ''i+1'' and the β conformation for residue ''i+2''; a niche4 has the α conformation for residues ''i+1'' and ''i+2'' and the β conformation for residue ''i+3''.
A niche occurs commonly at the
Niches are of two kinds, distinguished as niche3 (3 residues, ''i'' to ''i+2'') and niche4 (4 residues, ''i'' to ''i+3''). In a niche3 motif the δ+-binding carbonyl groups are from residues ''i'' and ''i+2'' while in a niche4 motif they are from residues ''i'' and ''i+3''.
A niche3 has the α conformation for residue ''i+1'' and the β conformation for residue ''i+2''; a niche4 has the α conformation for residues ''i+1'' and ''i+2'' and the β conformation for residue ''i+3''.
A niche occurs commonly at the C-terminus
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, carboxy tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein
Proteins are large biomolecules and macromolecules that comp ...
of α-helices
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix).
The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of l ...
and especially of 310 helices.
Metal ions that occur bound to niches in proteins are Na+, K+, Ca2+, and Mg2+. Proteins with regulatory cations often employ niches for metal binding (e.g., thrombin
Prothrombin (coagulation factor II) is encoded in the human by the F2-gene. It is proteolytically cleaved during the clotting process by the prothrombinase enzyme complex to form thrombin.
Thrombin (Factor IIa) (, fibrose, thrombase, throm ...
, Na+; annexin
Annexin is a common name for a group of cellular proteins. They are mostly found in eukaryotic organisms (animals, plants and fungi).
In humans, the annexins are found inside the cell. However some annexins (Annexin A1, Annexin A2, and Annexin A ...
, Ca2+; pyruvate dehydrogenase
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.
Pyruvate dehydrogenase is ...
, K+).
A major cation transporter in cells is calcium ATPase. In the Ca2+-bound crystal structures the two calcium ions side-by-side within the transmembrane domain
A transmembrane domain (TMD, TM domain) is a membrane-spanning protein domain. TMDs may consist of one or several alpha-helices or a transmembrane beta barrel. Because the interior of the lipid bilayer is hydrophobic, the amino acid residues in ...
are thought to be at the halfway stage of being transported. As well as being bound by various side chain carbonyl groups, one of these calcium ions is bound by a niche3/niche4 (both in the one motif) at residues 304–307 at the C-terminus of an α-helix.
A lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
side chain in the nuclear export receptor CRM1 is recognised specifically by a niche conformation that has to be adopted as a key part of the nuclear export signal
A nuclear export signal (NES) is a short target peptide containing 4 hydrophobic residues in a protein that targets it for export from the cell nucleus to the cytoplasm through the nuclear pore complex using nuclear transport. It has the opposit ...
of proteins exiting the nucleus.
A sodium ion in the Fluc fluoride channel is situated at the dyad axis of the dimer, bound tetrahedrally by two niche4s, one from each subunit. A Sodium ion bound in similar ways at the domain interface is seen in several other Na+-coupled transporters.
The Hsp70
The 70 kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat shock proteins. Proteins with similar structure exist in virtually all living organisms and play crucial roles in the development of can ...
interdomain linker region of 10 residues enables allosteric communication between two folded domains. The N-terminal part of the linker has a niche4 structure that is water-bound.
In the scorpion toxin BeM9 the side chain of arginine 60 binds the carbonyls of residues 61 and 63 as a niche3. The motif, loss of which alters the specificity of the protein for voltage-gated sodium channels, is named "arginine hand". The slightly unusual dihedral angles for a niche3 are because this niche3 accommodates two separate NH groups from the arginine's guanidino group.
Another small tripeptide motif that binds cations or δ+ groups via main-chain CO groups is called the catgrip.
References
External links
* Motivated Proteins* PDBeMotif
{{cite journal , last=Golovin , first=A , author2=Henrick , year=2008 , title=MSDmotif: exploring protein sites and motifs , journal=BMC Bioinformatics , volume=9 , issue=1 , pages=312 , doi=10.1186/1471-2105-9-312 , pmc=2491636 , pmid=18637174 , doi-access=free Protein structural motifs