Metagenome on:
[Wikipedia]
[Google]
[Amazon]
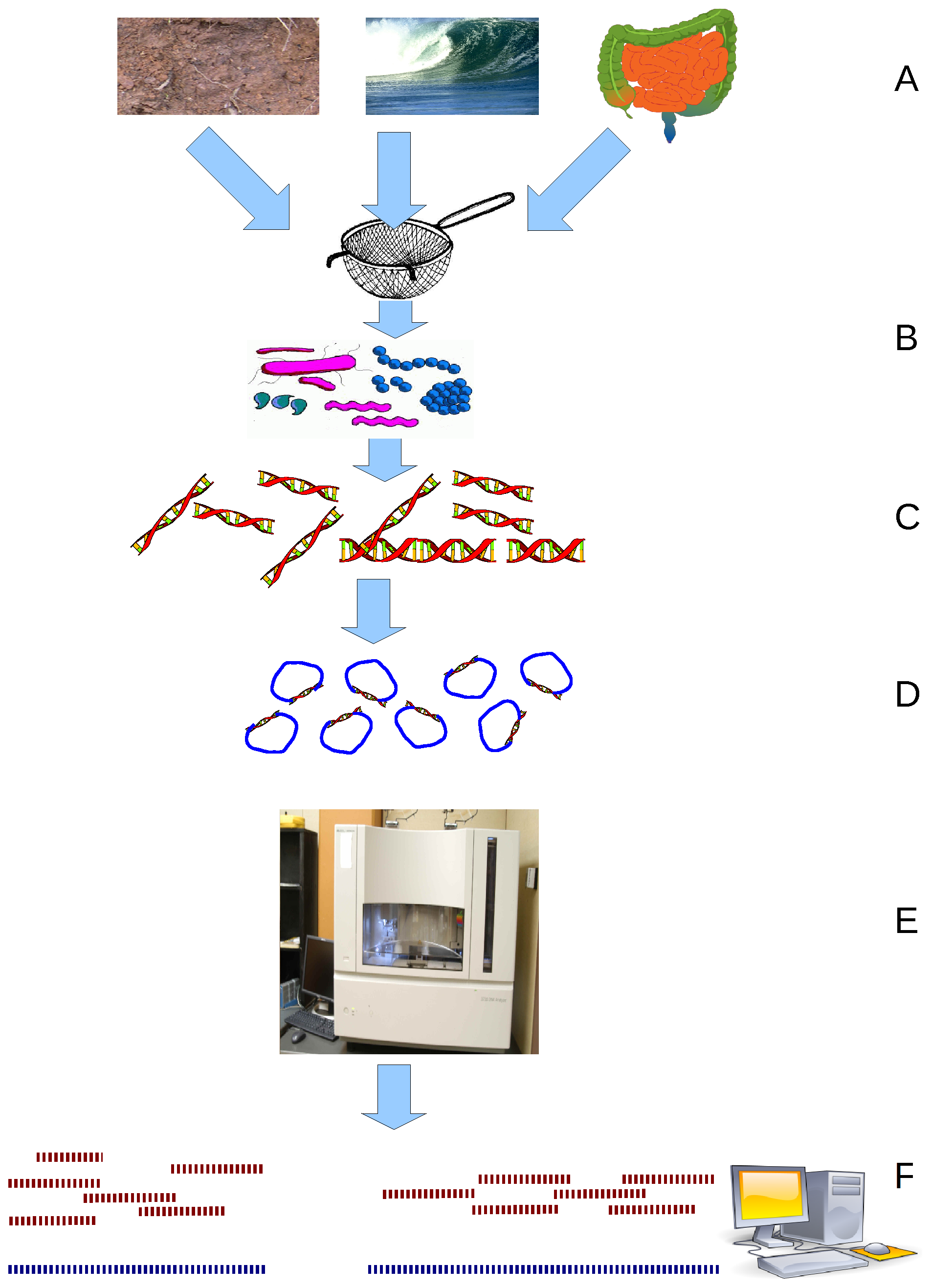 Metagenomics is the study of all genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the microbial composition of environmental and clinical samples without the need for time-consuming
Metagenomics is the study of all genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the microbial composition of environmental and clinical samples without the need for time-consuming
 The data generated by metagenomics experiments are both enormous and inherently noisy, containing fragmented data representing as many as 10,000 species. The sequencing of the cow
The data generated by metagenomics experiments are both enormous and inherently noisy, containing fragmented data representing as many as 10,000 species. The sequencing of the cow
and
mOTUs
ref name="sunagawa2013" /> and MetaPhyler, use universal marker genes to profile prokaryotic species. With th
mOTUs profiler
is possible to profile species without a reference genome, improving the estimation of microbial community diversity. Recent methods, such a
SLIMM
use read coverage landscape of individual reference genomes to minimize false-positive hits and get reliable relative abundances. In composition based binning, methods use intrinsic features of the sequence, such as oligonucleotide frequencies or codon usage bias. Once sequences are binned, it is possible to carry out comparative analysis of diversity and richness. After binning, assembled contigs are collected into "bins" each representing a species-like collection of organisms (see:
 Metagenomics can provide valuable insights into the functional ecology of environmental communities. Metagenomic analysis of the bacterial consortia found in the defecations of Australian sea lions suggests that nutrient-rich sea lion faeces may be an important nutrient source for coastal ecosystems. This is because the bacteria that are expelled simultaneously with the defecations are adept at breaking down the nutrients in the faeces into a bioavailable form that can be taken up into the food chain.
DNA sequencing can also be used more broadly to identify species present in a body of water, debris filtered from the air, sample of dirt, or animal's faeces, and even detect diet items from blood meals. This can establish the range of
Metagenomics can provide valuable insights into the functional ecology of environmental communities. Metagenomic analysis of the bacterial consortia found in the defecations of Australian sea lions suggests that nutrient-rich sea lion faeces may be an important nutrient source for coastal ecosystems. This is because the bacteria that are expelled simultaneously with the defecations are adept at breaking down the nutrients in the faeces into a bioavailable form that can be taken up into the food chain.
DNA sequencing can also be used more broadly to identify species present in a body of water, debris filtered from the air, sample of dirt, or animal's faeces, and even detect diet items from blood meals. This can establish the range of
Focus on Metagenomics
at '' Nature Reviews Microbiology'' journal website
The “Critical Assessment of Metagenome Interpretation” (CAMI) initiative
to evaluate methods in metagenomics {{Portal bar, Biology, Medicine Bioinformatics Genomics Environmental microbiology Microbiology techniques
 Metagenomics is the study of all genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the microbial composition of environmental and clinical samples without the need for time-consuming
Metagenomics is the study of all genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the microbial composition of environmental and clinical samples without the need for time-consuming culture
Culture ( ) is a concept that encompasses the social behavior, institutions, and Social norm, norms found in human societies, as well as the knowledge, beliefs, arts, laws, Social norm, customs, capabilities, Attitude (psychology), attitudes ...
of individual species.
Metagenomics has transformed microbial ecology
Microbial ecology (or environmental microbiology) is a discipline where the interaction of Microorganism, microorganisms and their environment are studied. Microorganisms are known to have important and harmful ecological relationships within t ...
and evolutionary biology
Evolutionary biology is the subfield of biology that studies the evolutionary processes such as natural selection, common descent, and speciation that produced the diversity of life on Earth. In the 1930s, the discipline of evolutionary biolo ...
by uncovering previously hidden biodiversity and metabolic capabilities. As the cost of DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
continues to decline, metagenomic studies now routinely profile hundreds to thousands of samples, enabling large-scale exploration of microbial communities and their roles in health and global ecosystems.
Metagenomic studies most commonly employ shotgun sequencing though long-read sequencing is being increasingly utilised as technologies advance. The field is also referred to as environmental genomics, ecogenomics, community genomics, or microbiomics and has significantly expanded the understanding of microbial life beyond what traditional cultivation-based methods can reveal.
Metagenomics is distinct from Amplicon sequencing, also referred to as Metabarcoding
Metabarcoding is the DNA barcoding, barcoding of DNA/RNA (or Environmental DNA, eDNA/Environmental DNA, eRNA) in a manner that allows for the simultaneous identification of many taxa within the same sample. The main difference between barcodin ...
or PCR-based sequencing. The main difference is the underlying methodology, since metagenomics targets all DNA in a sample, while Amplicon sequencing amplifies and sequences one or multiple specific genes. Data utilisation also differes between these two approaches. Amplicon sequencing provides mainly community profiles detailing which taxa are present in an sample, whereas metagenomics also recovers encoded enzymes and pathways. Amplicon sequencing was frequently used in early environmental gene sequencing focused on assessing specific highly conserved marker genes, such as the 16S rRNA gene, to profile microbial diversity. These studies demonstrated that the vast majority of microbial biodiversity had been missed by cultivation-based methods.
Etymology
The term "metagenomics" was first used by Jo Handelsman, Robert M. Goodman, Michelle R. Rondon, Jon Clardy, and Sean F. Brady, and first appeared in publication in 1998. The term metagenome referenced the idea that a collection of genes sequenced from the environment could be analyzed in a way analogous to the study of a singlegenome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
. In 2005, Kevin Chen and Lior Pachter (researchers at the University of California, Berkeley
The University of California, Berkeley (UC Berkeley, Berkeley, Cal, or California), is a Public university, public Land-grant university, land-grant research university in Berkeley, California, United States. Founded in 1868 and named after t ...
) defined metagenomics as "the application of modern genomics technique without the need for isolation and lab cultivation of individual species".
History
Conventionalsequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succ ...
begins with a culture of identical cells as a source of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
. However, early metagenomic studies revealed that there are probably large groups of microorganisms in many environments that cannot be cultured and thus cannot be sequenced. These early studies focused on 16S ribosomal RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
(rRNA) sequences which are relatively short, often conserved within a species, and generally different between species. Many 16S rRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal ...
sequences have been found which do not belong to any known cultured species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ...
, indicating that there are numerous non-isolated organisms. These surveys of ribosomal RNA genes taken directly from the environment revealed that cultivation based methods find less than 1% of the bacterial and archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
l species in a sample. Much of the interest in metagenomics comes from these discoveries that showed that the vast majority of microorganisms had previously gone unnoticed.
In the 1980s early molecular work in the field was conducted by Norman R. Pace and colleagues, who used PCR to explore the diversity of ribosomal RNA sequences. The insights gained from these breakthrough studies led Pace to propose the idea of cloning DNA directly from environmental samples as early as 1985. This led to the first report of isolating and cloning
Cloning is the process of producing individual organisms with identical genomes, either by natural or artificial means. In nature, some organisms produce clones through asexual reproduction; this reproduction of an organism by itself without ...
bulk DNA from an environmental sample, published by Pace and colleagues in 1991 while Pace was in the Department of Biology at Indiana University
Indiana University (IU) is a state university system, system of Public university, public universities in the U.S. state of Indiana. The system has two core campuses, five regional campuses, and two regional centers under the administration o ...
. Considerable efforts ensured that these were not PCR false positives and supported the existence of a complex community of unexplored species. Although this methodology was limited to exploring highly conserved, non-protein coding genes, it did support early microbial morphology-based observations that diversity was far more complex than was known by culturing methods. Soon after that in 1995, Healy reported the metagenomic isolation of functional genes from "zoolibraries" constructed from a complex culture of environmental organisms grown in the laboratory on dried grass
Poaceae ( ), also called Gramineae ( ), is a large and nearly ubiquitous family (biology), family of monocotyledonous flowering plants commonly known as grasses. It includes the cereal grasses, bamboos, the grasses of natural grassland and spe ...
es. After leaving the Pace laboratory, Edward DeLong continued in the field and has published work that has largely laid the groundwork for environmental phylogenies based on signature 16S sequences, beginning with his group's construction of libraries from marine samples.
In 2002, Mya Breitbart, Forest Rohwer, and colleagues used environmental shotgun sequencing (see below) to show that 200 liters of seawater contains over 5000 different viruses. Subsequent studies showed that there are more than a thousand viral species in human stool and possibly a million different viruses per kilogram of marine sediment
Marine sediment, or ocean sediment, or seafloor sediment, are deposits of insoluble particles that have accumulated on the seafloor. These particles either have their origins in soil and Rock (geology), rocks and have been Sediment transport, ...
, including many bacteriophages. Essentially all of the viruses in these studies were new species. In 2004, Gene Tyson, Jill Banfield, and colleagues at the University of California, Berkeley
The University of California, Berkeley (UC Berkeley, Berkeley, Cal, or California), is a Public university, public Land-grant university, land-grant research university in Berkeley, California, United States. Founded in 1868 and named after t ...
and the Joint Genome Institute sequenced DNA extracted from an acid mine drainage
Acid mine drainage, acid and metalliferous drainage (AMD), or acid rock drainage (ARD) is the outflow of acidic water from metal mines and coal mines.
Acid rock drainage occurs naturally within some environments as part of the rock weatherin ...
system. This effort resulted in the complete, or nearly complete, genomes for a handful of bacteria and archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
that had previously resisted attempts to culture them.
Beginning in 2003, Craig Venter, leader of the privately funded parallel of the Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
, has led the Global Ocean Sampling Expedition (GOS), circumnavigating the globe and collecting metagenomic samples throughout the journey. All of these samples were sequenced using shotgun sequencing, in hopes that new genomes (and therefore new organisms) would be identified. The pilot project, conducted in the Sargasso Sea, found DNA from nearly 2000 different species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ...
, including 148 types of bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
never before seen. Venter thoroughly explored the West Coast of the United States
The West Coast of the United States, also known as the Pacific Coast and the Western Seaboard, is the coastline along which the Western United States meets the North Pacific Ocean. The term typically refers to the Contiguous United States, contig ...
, and completed a two-year expedition in 2006 to explore the Baltic, Mediterranean
The Mediterranean Sea ( ) is a sea connected to the Atlantic Ocean, surrounded by the Mediterranean basin and almost completely enclosed by land: on the east by the Levant in West Asia, on the north by Anatolia in West Asia and Southern ...
, and Black
Black is a color that results from the absence or complete absorption of visible light. It is an achromatic color, without chroma, like white and grey. It is often used symbolically or figuratively to represent darkness.Eva Heller, ''P ...
Seas. Analysis of the metagenomic data collected during this journey revealed two groups of organisms, one composed of taxa adapted to environmental conditions of 'feast or famine', and a second composed of relatively fewer but more abundantly and widely distributed taxa primarily composed of plankton
Plankton are the diverse collection of organisms that drift in Hydrosphere, water (or atmosphere, air) but are unable to actively propel themselves against ocean current, currents (or wind). The individual organisms constituting plankton are ca ...
.
In 2005 Stephan C. Schuster at Penn State University
The Pennsylvania State University (Penn State or PSU) is a Public university, public Commonwealth System of Higher Education, state-related Land-grant university, land-grant research university with campuses and facilities throughout Pennsyl ...
and colleagues published the first sequences of an environmental sample generated with high-throughput sequencing, in this case massively parallel pyrosequencing
Pyrosequencing is a method of DNA sequencing (determining the order of nucleotides in DNA) based on the "sequencing by synthesis" principle, in which the sequencing is performed by detecting the nucleotide incorporated by a DNA polymerase. Pyrosequ ...
developed by 454 Life Sciences. Another early paper in this area appeared in 2006 by Robert Edwards, Forest Rohwer, and colleagues at San Diego State University
San Diego State University (SDSU) is a Public university, public research university in San Diego, California, United States. Founded in 1897, it is the third-oldest university and southernmost in the 23-member California State University (CS ...
.
there are few metagenomics laboratories in the world; for example, in the UK only the metagenomics labs at Great Ormond Street Hospital is recognised to carry out these tests. The cost is high (quoted as £1,300 in 2025), but this is expected to drop as the technology is developed.
Sequencing
Recovery of DNA sequences longer than a few thousandbase pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s from environmental samples was very difficult until recent advances in molecular biological techniques allowed the construction of libraries
A library is a collection of Book, books, and possibly other Document, materials and Media (communication), media, that is accessible for use by its members and members of allied institutions. Libraries provide physical (hard copies) or electron ...
in bacterial artificial chromosome
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid (or F-plasmid), used for transforming and cloning in bacteria, usually '' E. coli''. F-plasmids play a crucial role because they contain partiti ...
s (BACs), which provided better vectors for molecular cloning
Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their DNA replication, replication within Host (biology), host organisms. The use of the word ''cloning'' re ...
.
Shotgun metagenomics
Advances inbioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
, refinements of DNA amplification, and the proliferation of computational power have greatly aided the analysis of DNA sequences recovered from environmental samples, allowing the adaptation of shotgun sequencing to metagenomic samples (known also as whole metagenome shotgun or WMGS sequencing). The approach, used to sequence many cultured microorganisms and the human genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as the DNA within each of the 23 distinct chromosomes in the cell nucleus. A small DNA molecule is found within individual Mitochondrial DNA, mitochondria. These ar ...
, randomly shears DNA, sequences many short sequences, and reconstructs them into a consensus sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated sequence of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It represents the result ...
. Shotgun sequencing reveals genes present in environmental samples. Historically, clone libraries were used to facilitate this sequencing. However, with advances in high throughput sequencing technologies, the cloning step is no longer necessary and greater yields of sequencing data can be obtained without this labour-intensive bottleneck step. Shotgun metagenomics provides information both about which organisms are present and what metabolic processes are possible in the community. Because the collection of DNA from an environment is largely uncontrolled, the most abundant organisms in an environmental sample are most highly represented in the resulting sequence data. To achieve the high coverage needed to fully resolve the genomes of under-represented community members, large samples, often prohibitively so, are needed. On the other hand, the random nature of shotgun sequencing ensures that many of these organisms, which would otherwise go unnoticed using traditional culturing techniques, will be represented by at least some small sequence segments.
High-throughput sequencing
An advantage to high throughput sequencing is that this technique does not require cloning the DNA before sequencing, removing one of the main biases and bottlenecks in environmental sampling. The first metagenomic studies conducted using high-throughput sequencing used massively parallel 454 pyrosequencing. Three other technologies commonly applied to environmental sampling are the Ion Torrent Personal Genome Machine, the Illumina MiSeq or HiSeq and the Applied Biosystems SOLiD system. These techniques for sequencing DNA generate shorter fragments thanSanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
; Ion Torrent PGM System and 454 pyrosequencing typically produces ~400 bp reads, Illumina MiSeq produces 400-700bp reads (depending on whether paired end options are used), and SOLiD produce 25–75 bp reads. Historically, these read lengths were significantly shorter than the typical Sanger sequencing read length of ~750 bp, however the Illumina technology is quickly coming close to this benchmark. However, this limitation is compensated for by the much larger number of sequence reads. In 2009, pyrosequenced metagenomes generate 200–500 megabases, and Illumina platforms generate around 20–50 gigabases, but these outputs have increased by orders of magnitude in recent years.
An emerging approach combines shotgun sequencing and chromosome conformation capture (Hi-C), which measures the proximity of any two DNA sequences within the same cell, to guide microbial genome assembly. Long read sequencing technologies, including PacBio RSII and PacBio Sequel by Pacific Biosciences, and Nanopore MinION, GridION, PromethION by Oxford Nanopore Technologies, is another choice to get long shotgun sequencing reads that should make ease in assembling process.
Sequencing depth
An important consideration when sequencing for metagenomics is sequencing depth, the number of times each base is read by the sequencer; it can be thought of as resolution. The higher the sequencing depth, the larger the resultant file and number of contigs, and the higher the number of microbial genomes recovered. Higher depth metagenomes have been shown to have exceptionally high genome recovery, with tremendous novelty being reported. Low depth metagenomes have lower resolution on every taxonomic level than high depth samples.Bioinformatics
 The data generated by metagenomics experiments are both enormous and inherently noisy, containing fragmented data representing as many as 10,000 species. The sequencing of the cow
The data generated by metagenomics experiments are both enormous and inherently noisy, containing fragmented data representing as many as 10,000 species. The sequencing of the cow rumen
The rumen, also known as a paunch, is the largest stomach compartment in ruminants. The rumen and the reticulum make up the reticulorumen in ruminant animals. The diverse microbial communities in the rumen allows it to serve as the primary si ...
metagenome generated 279 gigabases, or 279 billion base pairs of nucleotide sequence data, while the human gut microbiome
A microbiome () is the community of microorganisms that can usually be found living together in any given habitat. It was defined more precisely in 1988 by Whipps ''et al.'' as "a characteristic microbial community occupying a reasonably wel ...
gene catalog identified 3.3 million genes assembled from 567.7 gigabases of sequence data. Collecting, curating, and extracting useful biological information from datasets of this size represent significant computational challenges for researchers.
Sequence pre-filtering
The first step of metagenomic data analysis requires the execution of certain pre-filtering steps, including the removal of redundant, low-quality sequences and sequences of probableeukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
origin (especially in metagenomes of human origin). The methods available for the removal of contaminating eukaryotic genomic DNA sequences include Eu-Detect and DeConseq.
Assembly
DNA sequence data from genomic and metagenomic projects are essentially the same, but genomic sequence data offers higher coverage while metagenomic data is usually highly non-redundant. Furthermore, the increased use of second-generation sequencing technologies with short read lengths means that much of future metagenomic data will be error-prone. Taken in combination, these factors make the assembly of metagenomic sequence reads into genomes difficult and unreliable. Misassemblies are caused by the presence of repetitive DNA sequences that make assembly especially difficult because of the difference in the relative abundance of species present in the sample. Misassemblies can also involve the combination of sequences from more than one species into chimeric contigs. There are several assembly programs, most of which can use information from paired-end tags in order to improve the accuracy of assemblies. Some programs, such as Phrap or Celera Assembler, were designed to be used to assemble singlegenome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
s but nevertheless produce good results when assembling metagenomic data sets. Other programs, such as Velvet assembler, have been optimized for the shorter reads produced by second-generation sequencing through the use of de Bruijn graphs. The use of reference genomes allows researchers to improve the assembly of the most abundant microbial species, but this approach is limited by the small subset of microbial phyla for which sequenced genomes are available. After an assembly is created, an additional challenge is "metagenomic deconvolution", or determining which sequences come from which species in the sample.
Species diversity
Gene annotations provide the "what", while measurements of species diversity provide the "who". In order to connect community composition and function in metagenomes, sequences must be binned. '' Binning'' is the process of associating a particular sequence with an organism. In similarity-based binning, methods such as BLAST are used to rapidly search for phylogenetic markers or otherwise similar sequences in existing public databases. This approach is implemented inMEGAN
Megan is a Welsh feminine given name, originally a diminutive form of Margaret. Margaret is from the Greek μαργαρίτης (''margarítēs''), Latin ''margarīta'', "pearl". Megan is one of the most popular Welsh-language names for women in ...
. Another tool, PhymmBL, uses interpolated Markov models to assign reads.MetaPhlAnand
AMPHORA
An amphora (; ; English ) is a type of container with a pointed bottom and characteristic shape and size which fit tightly (and therefore safely) against each other in storage rooms and packages, tied together with rope and delivered by land ...
are methods based on unique clade-specific markers for estimating organismal relative abundances with improved computational performances. Other tools, likmOTUs
ref name="sunagawa2013" /> and MetaPhyler, use universal marker genes to profile prokaryotic species. With th
mOTUs profiler
is possible to profile species without a reference genome, improving the estimation of microbial community diversity. Recent methods, such a
SLIMM
use read coverage landscape of individual reference genomes to minimize false-positive hits and get reliable relative abundances. In composition based binning, methods use intrinsic features of the sequence, such as oligonucleotide frequencies or codon usage bias. Once sequences are binned, it is possible to carry out comparative analysis of diversity and richness. After binning, assembled contigs are collected into "bins" each representing a species-like collection of organisms (see:
operational taxonomic unit
An operational taxonomic unit (OTU) is an operational definition used to classify groups of closely related individuals. The term was originally introduced in 1963 by Robert R. Sokal and Peter H. A. Sneath in the context of numerical taxonomy, wh ...
), to the best ability of the binning tool. Each bin consists of a ''metagenome-assembled genome'' (MAG), as all included sequences can be thought of being derived from the genome of the organism being represented. Tools based on single-copy genes such as CheckM and BUSCO can then be used to estimate the completeness percentage and contamination percentage of the MAG.
Gene prediction
Metagenomic analysis pipelines use two approaches in the annotation of coding regions in the assembled contigs. The first approach is to identify genes based upon homology with genes that are already publicly available insequence database
In the field of bioinformatics, a sequence database is a type of biological database that is composed of a large collection of computerized ("Digital data, digital") nucleic acid sequences, protein sequences, or other polymer sequences stored on a ...
s, usually by BLAST searches. This type of approach is implemented in the program MEGAN
Megan is a Welsh feminine given name, originally a diminutive form of Margaret. Margaret is from the Greek μαργαρίτης (''margarítēs''), Latin ''margarīta'', "pearl". Megan is one of the most popular Welsh-language names for women in ...
4. The second, '' ab initio'', uses intrinsic features of the sequence to predict coding regions based upon gene training sets from related organisms. This is the approach taken by programs such as GeneMark and GLIMMER. The main advantage of ''ab initio'' prediction is that it enables the detection of coding regions that lack homologs in the sequence databases; however, it is most accurate when there are large regions of contiguous genomic DNA available for comparison. Gene prediction is usually done after binning.
Comparative metagenomics
Comparative metagenomics involves analyzing differences in the taxonomic and functional composition of microbial communities across multiple samples or conditions. When focusing on features that vary between groups, this is often referred to as differential abundance analysis. Comparative analyses of metagenomes provide insights into how microbial communities vary across environments or hosts, helping to link community structure and function to ecological or health-related outcomes. Metagenomes are most commonly compared by analyzing taxonomic composition—such as differences in normalized species or genus abundance between groups— or taxonomicdiversity
Diversity, diversify, or diverse may refer to:
Business
*Diversity (business), the inclusion of people of different identities (ethnicity, gender, age) in the workforce
*Diversity marketing, marketing communication targeting diverse customers
* ...
, but can also be compared by sequence features such as k-mer profiles. Metadata on the environmental context of the metagenomic sample is important in comparative analyses, as it provides researchers with the ability to study the effect of habitat upon community structure and function.
Functional comparisons between metagenomes often involve profiling gene families or pathways using tools like HUMAnN3 or gutSMASH, which map reads to reference databases (e.g. KEGG, COG) or detect biosynthetic/metabolic gene clusters, enabling statistical comparison of functional potential across samples. This gene-centric approach emphasizes the functional complement of the community as a whole rather than taxonomic groups, and shows that the functional complements are analogous under similar environmental conditions.
Data analysis
Community metabolism
In many bacterial communities, natural or engineered (such asbioreactor
A bioreactor is any manufactured device or system that supports a biologically active environment. In one case, a bioreactor is a vessel in which a chemical reaction, chemical process is carried out which involves organisms or biochemistry, biochem ...
s), there is significant division of labor in metabolism ( syntrophy), during which the waste products of some organisms are metabolites for others. In one such system, the methanogen
Methanogens are anaerobic archaea that produce methane as a byproduct of their energy metabolism, i.e., catabolism. Methane production, or methanogenesis, is the only biochemical pathway for Adenosine triphosphate, ATP generation in methanogens. A ...
ic bioreactor, functional stability requires the presence of several syntrophic species ( Syntrophobacterales and Synergistia) working together in order to turn raw resources into fully metabolized waste (methane
Methane ( , ) is a chemical compound with the chemical formula (one carbon atom bonded to four hydrogen atoms). It is a group-14 hydride, the simplest alkane, and the main constituent of natural gas. The abundance of methane on Earth makes ...
). Using comparative gene studies and expression experiments with microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
s or proteomics
Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replicatio ...
researchers can piece together a metabolic network that goes beyond species boundaries. Such studies require detailed knowledge about which versions of which proteins are coded by which species and even by which strains of which species. Therefore, community genomic information is another fundamental tool (with metabolomics and proteomics) in the quest to determine how metabolites are transferred and transformed by a community.
Metatranscriptomics
Metagenomics allows researchers to access the functional and metabolic diversity of microbial communities, but it cannot show which of these processes are active. The extraction and analysis of metagenomicmRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
(the metatranscriptome) provides information on the regulation
Regulation is the management of complex systems according to a set of rules and trends. In systems theory, these types of rules exist in various fields of biology and society, but the term has slightly different meanings according to context. Fo ...
and expression profiles of complex communities. Because of the technical difficulties (the short half-life of mRNA, for example) in the collection of environmental RNA there have been relatively few ''in situ
is a Latin phrase meaning 'in place' or 'on site', derived from ' ('in') and ' ( ablative of ''situs'', ). The term typically refers to the examination or occurrence of a process within its original context, without relocation. The term is use ...
'' metatranscriptomic studies of microbial communities to date. While originally limited to microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrate—usu ...
technology, metatranscriptomics studies have made use of transcriptomics technologies to measure whole-genome expression and quantification of a microbial community, first employed in analysis of ammonia oxidation in soils.
Viruses
Metagenomic sequencing is particularly useful in the study of viral communities. As viruses lack a shared universal phylogenetic marker (as 16S RNA for bacteria and archaea, and 18S RNA for eukarya), the only way to access the genetic diversity of the viral community from an environmental sample is through metagenomics. Viral metagenomes (also called viromes) should thus provide more and more information about viral diversity and evolution. For example, a metagenomic pipeline called Giant Virus Finder showed the first evidence of existence of giant viruses in a saline desert and in Antarctic dry valleys.Applications
Metagenomics has the potential to advance knowledge in a wide variety of fields. It can also be applied to solve practical challenges inmedicine
Medicine is the science and Praxis (process), practice of caring for patients, managing the Medical diagnosis, diagnosis, prognosis, Preventive medicine, prevention, therapy, treatment, Palliative care, palliation of their injury or disease, ...
, engineering
Engineering is the practice of using natural science, mathematics, and the engineering design process to Problem solving#Engineering, solve problems within technology, increase efficiency and productivity, and improve Systems engineering, s ...
, agriculture
Agriculture encompasses crop and livestock production, aquaculture, and forestry for food and non-food products. Agriculture was a key factor in the rise of sedentary human civilization, whereby farming of domesticated species created ...
, sustainability
Sustainability is a social goal for people to co-exist on Earth over a long period of time. Definitions of this term are disputed and have varied with literature, context, and time. Sustainability usually has three dimensions (or pillars): env ...
and ecology
Ecology () is the natural science of the relationships among living organisms and their Natural environment, environment. Ecology considers organisms at the individual, population, community (ecology), community, ecosystem, and biosphere lev ...
.
Agriculture
Thesoil
Soil, also commonly referred to as earth, is a mixture of organic matter, minerals, gases, water, and organisms that together support the life of plants and soil organisms. Some scientific definitions distinguish dirt from ''soil'' by re ...
s in which plants grow are inhabited by microbial communities, with one gram of soil containing around 109-1010 microbial cells which comprise about one gigabase of sequence information. The microbial communities which inhabit soils are some of the most complex known to science, and remain poorly understood despite their economic importance. Microbial consortia perform a wide variety of ecosystem services necessary for plant growth, including fixing atmospheric nitrogen, nutrient cycling
A nutrient cycle (or ecological recycling) is the movement and exchange of inorganic and organic matter back into the production of matter. Energy flow is a unidirectional and noncyclic pathway, whereas the movement of mineral nutrients is cyc ...
, disease suppression, and sequester iron
Iron is a chemical element; it has symbol Fe () and atomic number 26. It is a metal that belongs to the first transition series and group 8 of the periodic table. It is, by mass, the most common element on Earth, forming much of Earth's o ...
and other metal
A metal () is a material that, when polished or fractured, shows a lustrous appearance, and conducts electrical resistivity and conductivity, electricity and thermal conductivity, heat relatively well. These properties are all associated wit ...
s. Functional metagenomics strategies are being used to explore the interactions between plants and microbes through cultivation-independent study of these microbial communities. By allowing insights into the role of previously uncultivated or rare community members in nutrient cycling and the promotion of plant growth, metagenomic approaches can contribute to improved disease detection in crop
A crop is a plant that can be grown and harvested extensively for profit or subsistence. In other words, a crop is a plant or plant product that is grown for a specific purpose such as food, Fiber, fibre, or fuel.
When plants of the same spe ...
s and livestock
Livestock are the Domestication, domesticated animals that are raised in an Agriculture, agricultural setting to provide labour and produce diversified products for consumption such as meat, Egg as food, eggs, milk, fur, leather, and wool. The t ...
and the adaptation of enhanced farming
Agriculture encompasses crop and livestock production, aquaculture, and forestry for food and non-food products. Agriculture was a key factor in the rise of sedentary human civilization, whereby farming of domesticated species created ...
practices which improve crop health by harnessing the relationship between microbes and plants.
Biofuel
Biofuel
Biofuel is a fuel that is produced over a short time span from Biomass (energy), biomass, rather than by the very slow natural processes involved in the formation of fossil fuels such as oil. Biofuel can be produced from plants or from agricu ...
s are fuel
A fuel is any material that can be made to react with other substances so that it releases energy as thermal energy or to be used for work (physics), work. The concept was originally applied solely to those materials capable of releasing chem ...
s derived from biomass
Biomass is a term used in several contexts: in the context of ecology it means living organisms, and in the context of bioenergy it means matter from recently living (but now dead) organisms. In the latter context, there are variations in how ...
conversion, as in the conversion of cellulose
Cellulose is an organic compound with the chemical formula, formula , a polysaccharide consisting of a linear chain of several hundred to many thousands of glycosidic bond, β(1→4) linked glucose, D-glucose units. Cellulose is an important s ...
contained in corn
Maize (; ''Zea mays''), also known as corn in North American English, is a tall stout Poaceae, grass that produces cereal grain. It was domesticated by indigenous peoples of Mexico, indigenous peoples in southern Mexico about 9,000 years ago ...
stalks, switchgrass
''Panicum virgatum'', commonly known as switchgrass, is a perennial warm season bunchgrass native to North America, where it occurs naturally from 55th parallel north, 55°N latitude in Canada southwards into the United States and Mexico. Switch ...
, and other biomass into cellulosic ethanol
Cellulosic ethanol is ethanol (ethyl alcohol) produced from cellulose (the stringy fiber of a plant) rather than from the plant's seeds or fruit. It can be produced from grasses, wood, algae, or other plants. It is generally discussed for use as a ...
. This process is dependent upon microbial consortia (association) that transform the cellulose into sugar
Sugar is the generic name for sweet-tasting, soluble carbohydrates, many of which are used in food. Simple sugars, also called monosaccharides, include glucose
Glucose is a sugar with the Chemical formula#Molecular formula, molecul ...
s, followed by the fermentation
Fermentation is a type of anaerobic metabolism which harnesses the redox potential of the reactants to make adenosine triphosphate (ATP) and organic end products. Organic molecules, such as glucose or other sugars, are catabolized and reduce ...
of the sugars into ethanol
Ethanol (also called ethyl alcohol, grain alcohol, drinking alcohol, or simply alcohol) is an organic compound with the chemical formula . It is an Alcohol (chemistry), alcohol, with its formula also written as , or EtOH, where Et is the ps ...
. Microbes also produce a variety of sources of bioenergy
Bioenergy is a type of renewable energy that is derived from plants and animal waste. The Biomass (energy), biomass that is used as input materials consists of recently living (but now dead) organisms, mainly plants. Thus, Fossil fuel, fossil fu ...
including methane
Methane ( , ) is a chemical compound with the chemical formula (one carbon atom bonded to four hydrogen atoms). It is a group-14 hydride, the simplest alkane, and the main constituent of natural gas. The abundance of methane on Earth makes ...
and hydrogen
Hydrogen is a chemical element; it has chemical symbol, symbol H and atomic number 1. It is the lightest and abundance of the chemical elements, most abundant chemical element in the universe, constituting about 75% of all baryon, normal matter ...
.
The efficient industrial-scale deconstruction of biomass requires novel enzymes
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as pro ...
with higher productivity and lower cost. Metagenomic approaches to the analysis of complex microbial communities allow the targeted screening of enzymes
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as pro ...
with industrial applications in biofuel production, such as glycoside hydrolases. Furthermore, knowledge of how these microbial communities function is required to control them, and metagenomics is a key tool in their understanding. Metagenomic approaches allow comparative analyses between convergent microbial systems like biogas
Biogas is a gaseous renewable energy source produced from raw materials such as agricultural waste, manure, municipal waste, plant material, sewage, green waste, Wastewater treatment, wastewater, and food waste. Biogas is produced by anaerobic ...
fermenters or insect
Insects (from Latin ') are Hexapoda, hexapod invertebrates of the class (biology), class Insecta. They are the largest group within the arthropod phylum. Insects have a chitinous exoskeleton, a three-part body (Insect morphology#Head, head, ...
herbivore
A herbivore is an animal anatomically and physiologically evolved to feed on plants, especially upon vascular tissues such as foliage, fruits or seeds, as the main component of its diet. These more broadly also encompass animals that eat ...
s such as the fungus garden of the leafcutter ants.
Biotechnology
Microbial communities produce a vast array of biologically active chemicals that are used in competition and communication. Many of the drugs in use today were originally uncovered in microbes; recent progress in mining the rich genetic resource of non-culturable microbes has led to the discovery of new genes, enzymes, and natural products. The application of metagenomics has allowed the development ofcommodity
In economics, a commodity is an economic goods, good, usually a resource, that specifically has full or substantial fungibility: that is, the Market (economics), market treats instances of the good as equivalent or nearly so with no regard to w ...
and fine chemicals, agrochemicals and pharmaceuticals
Medication (also called medicament, medicine, pharmaceutical drug, medicinal product, medicinal drug or simply drug) is a drug used to diagnose, cure, treat, or prevent disease. Drug therapy ( pharmacotherapy) is an important part of the ...
where the benefit of enzyme-catalyzed chiral synthesis is increasingly recognized.
Two types of analysis are used in the bioprospecting of metagenomic data: function-driven screening for an expressed trait, and sequence-driven screening for DNA sequences of interest. Function-driven analysis seeks to identify clones expressing a desired trait or useful activity, followed by biochemical characterization and sequence analysis. This approach is limited by availability of a suitable screen and the requirement that the desired trait be expressed in the host cell. Moreover, the low rate of discovery (less than one per 1,000 clones screened) and its labor-intensive nature further limit this approach. In contrast, sequence-driven analysis uses conserved DNA sequences to design PCR primers to screen clones for the sequence of interest. In comparison to cloning-based approaches, using a sequence-only approach further reduces the amount of bench work required. The application of massively parallel sequencing also greatly increases the amount of sequence data generated, which require high-throughput bioinformatic analysis pipelines. The sequence-driven approach to screening is limited by the breadth and accuracy of gene functions present in public sequence databases. In practice, experiments make use of a combination of both functional and sequence-based approaches based upon the function of interest, the complexity of the sample to be screened, and other factors. An example of success using metagenomics as a biotechnology for drug discovery is illustrated with the malacidin antibiotics.
Ecology
 Metagenomics can provide valuable insights into the functional ecology of environmental communities. Metagenomic analysis of the bacterial consortia found in the defecations of Australian sea lions suggests that nutrient-rich sea lion faeces may be an important nutrient source for coastal ecosystems. This is because the bacteria that are expelled simultaneously with the defecations are adept at breaking down the nutrients in the faeces into a bioavailable form that can be taken up into the food chain.
DNA sequencing can also be used more broadly to identify species present in a body of water, debris filtered from the air, sample of dirt, or animal's faeces, and even detect diet items from blood meals. This can establish the range of
Metagenomics can provide valuable insights into the functional ecology of environmental communities. Metagenomic analysis of the bacterial consortia found in the defecations of Australian sea lions suggests that nutrient-rich sea lion faeces may be an important nutrient source for coastal ecosystems. This is because the bacteria that are expelled simultaneously with the defecations are adept at breaking down the nutrients in the faeces into a bioavailable form that can be taken up into the food chain.
DNA sequencing can also be used more broadly to identify species present in a body of water, debris filtered from the air, sample of dirt, or animal's faeces, and even detect diet items from blood meals. This can establish the range of invasive species
An invasive species is an introduced species that harms its new environment. Invasive species adversely affect habitats and bioregions, causing ecological, environmental, and/or economic damage. The term can also be used for native spec ...
and endangered species
An endangered species is a species that is very likely to become extinct in the near future, either worldwide or in a particular political jurisdiction. Endangered species may be at risk due to factors such as habitat loss, poaching, inv ...
, and track seasonal populations.
Environmental remediation
Metagenomics can improve strategies for monitoring the impact ofpollutant
A pollutant or novel entity is a substance or energy introduced into the environment that has undesired effect, or adversely affects the usefulness of a resource. These can be both naturally forming (i.e. minerals or extracted compounds like oi ...
s on ecosystem
An ecosystem (or ecological system) is a system formed by Organism, organisms in interaction with their Biophysical environment, environment. The Biotic material, biotic and abiotic components are linked together through nutrient cycles and en ...
s and for cleaning up contaminated environments. Increased understanding of how microbial communities cope with pollutants improves assessments of the potential of contaminated sites to recover from pollution and increases the chances of bioaugmentation or biostimulation trials to succeed.
Gut microbe characterization
Microbial communities play a key role in preserving humanhealth
Health has a variety of definitions, which have been used for different purposes over time. In general, it refers to physical and emotional well-being, especially that associated with normal functioning of the human body, absent of disease, p ...
, but their composition and the mechanism by which they do so remains mysterious. Metagenomic sequencing is being used to characterize the microbial communities from 15–18 body sites from at least 250 individuals. This is part of the Human Microbiome initiative with primary goals to determine if there is a core human microbiome, to understand the changes in the human microbiome that can be correlated with human health, and to develop new technological and bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
tools to support these goals.
Another medical study as part of the MetaHit (Metagenomics of the Human Intestinal Tract) project consisted of 124 individuals from Denmark and Spain consisting of healthy, overweight, and irritable bowel disease patients. The study attempted to categorize the depth and phylogenetic diversity of gastrointestinal bacteria. Using Illumina GA sequence data and SOAPdenovo, a de Bruijn graph-based tool specifically designed for assembly short reads, they were able to generate 6.58 million contigs greater than 500 bp for a total contig length of 10.3 Gb and a N50 length of 2.2 kb.
The study demonstrated that two bacterial divisions, Bacteroidetes and Firmicutes, constitute over 90% of the known phylogenetic categories that dominate distal gut bacteria. Using the relative gene frequencies found within the gut these researchers identified 1,244 metagenomic clusters that are critically important for the health of the intestinal tract. There are two types of functions in these range clusters: housekeeping and those specific to the intestine. The housekeeping gene clusters are required in all bacteria and are often major players in the main metabolic pathways including central carbon metabolism and amino acid synthesis. The gut-specific functions include adhesion to host proteins and the harvesting of sugars from globoseries glycolipids. Patients with irritable bowel syndrome were shown to exhibit 25% fewer genes and lower bacterial diversity than individuals not suffering from irritable bowel syndrome indicating that changes in patients' gut biome diversity may be associated with this condition.
While these studies highlight some potentially valuable medical applications, only 31–48.8% of the reads could be aligned to 194 public human gut bacterial genomes and 7.6–21.2% to bacterial genomes available in GenBank which indicates that there is still far more research necessary to capture novel bacterial genomes.
In the Human Microbiome Project (HMP), gut microbial communities were assayed using high-throughput DNA sequencing. HMP showed that, unlike individual microbial species, many metabolic processes were present among all body habitats with varying frequencies. Microbial communities of 649 metagenomes drawn from seven primary body sites on 102 individuals were studied as part of the human microbiome project. The metagenomic analysis revealed variations in niche specific abundance among 168 functional modules and 196 metabolic pathways within the microbiome. These included glycosaminoglycan degradation in the gut, as well as phosphate and amino acid transport linked to host phenotype (vaginal pH) in the posterior fornix. The HMP has brought to light the utility of metagenomics in diagnostics and evidence-based medicine
Evidence-based medicine (EBM) is "the conscientious, explicit and judicious use of current best evidence in making decisions about the care of individual patients. It means integrating individual clinical expertise with the best available exte ...
. Thus metagenomics is a powerful tool to address many of the pressing issues in the field of personalized medicine.
In animals, metagenomics can be used to profile their gut microbiomes and enable detection of antibiotic-resistant bacteria. This can have implications in monitoring the spread of diseases from wildlife to farmed animals and humans.
Infectious disease diagnosis
Differentiating between infectious and non-infectious illness, and identifying the underlying etiology of infection, can be challenging. For example, more than half of cases ofencephalitis
Encephalitis is inflammation of the Human brain, brain. The severity can be variable with symptoms including reduction or alteration in consciousness, aphasia, headache, fever, confusion, a stiff neck, and vomiting. Complications may include se ...
remain undiagnosed, despite extensive testing using state-of-the-art clinical laboratory methods. Clinical metagenomic sequencing shows promise as a sensitive and rapid method to diagnose infection by comparing genetic material found in a patient's sample to databases of all known microscopic human pathogens and thousands of other bacterial, viral, fungal, and parasitic organisms and databases on antimicrobial resistances gene sequences with associated clinical phenotypes.
Arbovirus surveillance
Metagenomics is helpful to characterize the diversity and ecology of viruses spread by hematophagous (blood-feeding) arthropods such as mosquitoes and ticks, calledarbovirus
Arbovirus is an informal name for any virus that is Transmission (medicine), transmitted by arthropod Vector (epidemiology), vectors. The term ''arbovirus'' is a portmanteau word (''ar''thropod-''bo''rne ''virus''). ''Tibovirus'' (''ti''ck-''bo ...
es. It can also be used as a tool by public health officials and organizations to surveil arboviruses in circulation in wild arthropod populations.
Dietary estimation
Metagenomic Estimation of Dietary Intake (MEDI), enables reconstruction of individual dietary profiles by detecting food-derived DNA in human stool metagenomes. MEDI has shown concordance with food frequency questionnaires, tracked dietary shifts in infants, and identified diet–health associations in large cohorts without dietary records.See also
* Binning * Epidemiology and sewage * Metaproteomics *Microbial ecology
Microbial ecology (or environmental microbiology) is a discipline where the interaction of Microorganism, microorganisms and their environment are studied. Microorganisms are known to have important and harmful ecological relationships within t ...
* Pathogenomics
* Virome analysis
References
External links
Focus on Metagenomics
at '' Nature Reviews Microbiology'' journal website
The “Critical Assessment of Metagenome Interpretation” (CAMI) initiative
to evaluate methods in metagenomics {{Portal bar, Biology, Medicine Bioinformatics Genomics Environmental microbiology Microbiology techniques