Genetic distance on:
[Wikipedia]
[Google]
[Amazon]
 Genetic distance is a measure of the genetic divergence between
Genetic distance is a measure of the genetic divergence between

 The Kimura two parameter model (K2P) was developed in 1980 by Japanese biologist Motoo Kimura. It is compatible with the neutral theory of evolution, which was also developed by the same author. As depicted in Figure 4, this measure of genetic distance accounts for the type of mutation occurring, namely whether it is a transition (i.e. purine to purine or pyrimidine to pyrimidine) or a transversion (i.e. purine to pyrimidine or vice versa). With this information, the following formula can be derived:
:
where P is and Q is , with being the number of transition type conversions, being the number of transversion type conversions, and being the number of nucleotides sites compared.
It is worth noting when transition and transversion type substitutions have an equal chance of occurring, and is assumed to equal , then the above formula can be reduced down to the Jukes Cantor model. In practice however, is typically larger than .
It has been shown that while K2P works well in classifying distantly-related species, it is not always the best choice for comparing closely-related species. In these cases, it may be better to use p-distance instead.
The Kimura two parameter model (K2P) was developed in 1980 by Japanese biologist Motoo Kimura. It is compatible with the neutral theory of evolution, which was also developed by the same author. As depicted in Figure 4, this measure of genetic distance accounts for the type of mutation occurring, namely whether it is a transition (i.e. purine to purine or pyrimidine to pyrimidine) or a transversion (i.e. purine to pyrimidine or vice versa). With this information, the following formula can be derived:
:
where P is and Q is , with being the number of transition type conversions, being the number of transversion type conversions, and being the number of nucleotides sites compared.
It is worth noting when transition and transversion type substitutions have an equal chance of occurring, and is assumed to equal , then the above formula can be reduced down to the Jukes Cantor model. In practice however, is typically larger than .
It has been shown that while K2P works well in classifying distantly-related species, it is not always the best choice for comparing closely-related species. In these cases, it may be better to use p-distance instead.
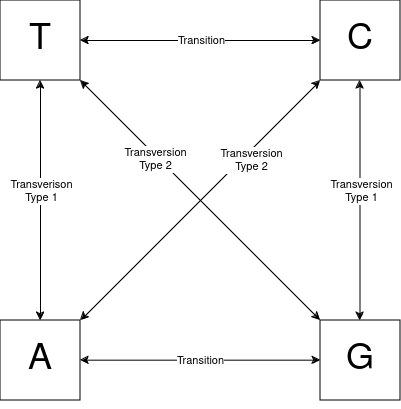 The Kimura three parameter (K3P) model was first published in 1981. This measure assumes three rates of substitution when nucleotides mutate, which can be seen in Figure 5. There is one rate for transition type mutations, one rate for transversion type mutations to corresponding bases (e.g. G to C; transversion type 1 in the figure), and one rate for transversion type mutations to non-corresponding bases (e.g. G to T; transversion type 2 in the figure).
With these rates of substitution, the following formula can be derived:
:
where is the probability of a transition type mutation, is the probability of a transversion type mutation to a corresponding base, and is the probability of a transversion type mutation to a non-corresponding base. When and are assumed to be equal, this reduces down to the Kimura 2 parameter distance.
The Kimura three parameter (K3P) model was first published in 1981. This measure assumes three rates of substitution when nucleotides mutate, which can be seen in Figure 5. There is one rate for transition type mutations, one rate for transversion type mutations to corresponding bases (e.g. G to C; transversion type 1 in the figure), and one rate for transversion type mutations to non-corresponding bases (e.g. G to T; transversion type 2 in the figure).
With these rates of substitution, the following formula can be derived:
:
where is the probability of a transition type mutation, is the probability of a transversion type mutation to a corresponding base, and is the probability of a transversion type mutation to a non-corresponding base. When and are assumed to be equal, this reduces down to the Kimura 2 parameter distance.
 Similar to Euclidean distance, Czekanowski distance involves calculated the distance between points of allele frequency that are graphed on an axis created by . However, Czekanowski assumes a direct path is not available and sums the sides of the triangle formed by the data points instead of finding the hypotenuse. This formula is nicknamed the Manhattan distance because its methodology is similar to the nature of the New York City burrow. Manhattan is mainly built on a grid system requiring resentence to only make 90 degree turns during travel, which parallels the thinking of the formula.
and : Allele frequencies at locus u in populations X and Y, respectively
and : X-axis value of the frequency of an allele for X and Y populations
and : Y-axis value of the frequency of an allele for X and Y populations
Similar to Euclidean distance, Czekanowski distance involves calculated the distance between points of allele frequency that are graphed on an axis created by . However, Czekanowski assumes a direct path is not available and sums the sides of the triangle formed by the data points instead of finding the hypotenuse. This formula is nicknamed the Manhattan distance because its methodology is similar to the nature of the New York City burrow. Manhattan is mainly built on a grid system requiring resentence to only make 90 degree turns during travel, which parallels the thinking of the formula.
and : Allele frequencies at locus u in populations X and Y, respectively
and : X-axis value of the frequency of an allele for X and Y populations
and : Y-axis value of the frequency of an allele for X and Y populations
 Similar to Czekanowski distance, Roger's distance involves calculating the distance between points of allele frequency. However, this method takes the direct distance between the points.
and : Allele frequencies at locus u in populations X and Y, respectively
: Total number of microsatallite loci examined
Similar to Czekanowski distance, Roger's distance involves calculating the distance between points of allele frequency. However, this method takes the direct distance between the points.
and : Allele frequencies at locus u in populations X and Y, respectively
: Total number of microsatallite loci examined
GENDIST
** Nei's standard genetic distance 1972 ** Cavalli-Sforza and Edwards 1967 ** Reynolds, Weir, and Cockerham's 1983
TFPGA
** Nei's standard genetic distance (original and unbiased) ** Nei's minimum genetic distance (original and unbiased) ** Wright's (1978) modification of Roger's (1972) distance ** Reynolds, Weir, and Cockerham's 1983
GDA
POPGENE
** Commonly used genetic distances and gene diversity analysis
** Nei's standard genetic distance 1972 ** Nei's DA distance between populations 1983
''The Estimation of Genetic Distance and Population Substructure from Microsatellite allele frequency data.'', Brent W. Murray (May 1996), McMaster University website on genetic distance
{{DEFAULTSORT:Genetic Distance Genetics concepts Phylogenetics
 Genetic distance is a measure of the genetic divergence between
Genetic distance is a measure of the genetic divergence between species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ...
or between populations within a species, whether the distance measures time from common ancestor
Common descent is a concept in evolutionary biology applicable when one species is the ancestor of two or more species later in time. According to modern evolutionary biology, all living beings could be descendants of a unique ancestor commonl ...
or degree of differentiation. Populations with many similar allele
An allele is a variant of the sequence of nucleotides at a particular location, or Locus (genetics), locus, on a DNA molecule.
Alleles can differ at a single position through Single-nucleotide polymorphism, single nucleotide polymorphisms (SNP), ...
s have small genetic distances. This indicates that they are closely related and have a recent common ancestor.
Genetic distance is useful for reconstructing the history of populations, such as the multiple human expansions out of Africa. It is also used for understanding the origin of biodiversity
Biodiversity is the variability of life, life on Earth. It can be measured on various levels. There is for example genetic variability, species diversity, ecosystem diversity and Phylogenetics, phylogenetic diversity. Diversity is not distribut ...
. For example, the genetic distances between different breeds of domesticated animals are often investigated in order to determine which breeds should be protected to maintain genetic diversity.
Biological foundation
Life on earth began from very simple unicellular organisms evolving into most complexmulticellular
A multicellular organism is an organism that consists of more than one cell (biology), cell, unlike unicellular organisms. All species of animals, Embryophyte, land plants and most fungi are multicellular, as are many algae, whereas a few organism ...
organisms through the course of over three billion years. Creating a comprehensive tree of life
The tree of life is a fundamental archetype in many of the world's mythology, mythological, religion, religious, and philosophy, philosophical traditions. It is closely related to the concept of the sacred tree.Giovino, Mariana (2007). ''The ...
that represents all the organisms that have ever lived on earth is important for understanding the evolution of life in the face of all challenges faced by living organisms to deal with similar challenges in future. Evolutionary biologists have attempted to create evolutionary or phylogenetic
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical dat ...
trees encompassing as many organisms as possible based on the available resources. Fossil
A fossil (from Classical Latin , ) is any preserved remains, impression, or trace of any once-living thing from a past geological age. Examples include bones, shells, exoskeletons, stone imprints of animals or microbes, objects preserve ...
dating
Dating is a stage of Romance (love), romantic relationships in which individuals engage in activity together, often with the intention of evaluating each other's suitability as a partner in a future intimate relationship. It falls into the cate ...
and molecular clock are the two means of generating evolutionary history of living organisms. Fossil record is random, incomplete and does not provide a continuous chain of events like a movie with missing frames cannot tell the whole plot of the movie.
Molecular clocks on the other hand are specific sequences of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
, RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
or protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s (amino acids) that are used to determine at molecular level the similarities and differences among species, to find out the timeline of divergence, and to trace back the common ancestor of species based on the mutation rate
In genetics, the mutation rate is the frequency of new mutations in a single gene, nucleotide sequence, or organism over time. Mutation rates are not constant and are not limited to a single type of mutation; there are many different types of mu ...
s and sequence changes being accumulated in those specific sequences. The primary driver of evolution is the mutation or changes in genes and accounting for those changes over time determines the approximate genetic distance between species. These specific molecular clocks are fairly conserved across a range of species and have a constant rate of mutation like a clock and are calibrated based on evolutionary events (fossil records). For example, gene for alpha-globin (constituent of hemoglobin) mutates at a rate of 0.56 per base pair per billion years. The molecular clock can fill those gaps created by missing fossil records.
In the genome of an organism
An organism is any life, living thing that functions as an individual. Such a definition raises more problems than it solves, not least because the concept of an individual is also difficult. Many criteria, few of them widely accepted, have be ...
, each gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
is located at a specific place called the locus for that gene. Allelic variations at these loci cause phenotypic variation within species (e.g. hair colour, eye colour). However, most alleles do not have an observable impact on the phenotype. Within a population new alleles generated by mutation either die out or spread throughout the population. When a population is split into different isolated populations (by either geographical or ecological factors), mutations that occur after the split will be present only in the isolated population. Random fluctuation of allele frequencies also produces genetic differentiation between populations. This process is known as genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
. By examining the differences between allele frequencies between the populations and computing genetic distance, we can estimate how long ago the two populations were separated.
Let’s suppose a sequence of DNA or a hypothetical gene that has mutation rate of one base per 10 million years. Using this sequence of DNA, the divergence of two different species or genetic distance between two different species can be determined by counting the number of base pair differences among them. For example, in Figure 2 a difference of 4 bases in the hypothetical sequence among those two species would indicate that they diverged 40 million years ago, and their common ancestor would have lived at least 20 million years ago before their divergence. Based on molecular clock, the equation below can be used to calculate the time since divergence.
''Number of mutation ÷ Mutation per year (rate of mutation) = time since divergence''

Process of determining genetic distance
Recent advancement in sequencing technology and the availability of comprehensive genomic databases and bioinformatics tools that are capable of storing and processing colossal amount of data generated by the advanced sequencing technology has tremendously improved evolutionary studies and the understanding of evolutionary relationships among species.Markers for genetic distance
Different biomolecular markers such DNA, RNA andamino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
sequences (protein) can be used for determining the genetic distance.
The selection criteria of appropriate biomarker for genetic distance entails the following three steps:
# choice of variability
# choice of specific region of DNA or RNA
# the use of technique
Technique or techniques may refer to:
Music
* The Techniques, a Jamaican rocksteady vocal group of the 1960s
* Technique (band), a British female synth pop band in the 1990s
* ''Technique'' (album), by New Order, 1989
* ''Techniques'' (album), by ...
The choice of variability depends on the intended outcome. For example, very high level of variability is recommended for demographic studies and parentage analyses, medium to high variability for comparing distinct populations, and moderate to very low variability is recommended for phylogenetic studies. The genomic localization and ploidy of the marker is also an important factor. For example, the gene copy number is inversely proportional to the robustness with haploid
Ploidy () is the number of complete sets of chromosomes in a cell (biology), cell, and hence the number of possible alleles for Autosome, autosomal and Pseudoautosomal region, pseudoautosomal genes. Here ''sets of chromosomes'' refers to the num ...
genome (mitochondrial DNA
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondrion, mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the D ...
) more prone to genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
than diploid genome (nuclear DNA
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. ...
).
The choice and examples of molecular markers for evolutionary biology studies.
Application of genetic distance
* Phylogenetics: Exploring the genetic distance among species can help in establishing evolutionary relationships among them, the time of divergence between them and creating a comprehensive phylogenetic tree that connect them to their common ancestors. * Accuracy of genomic prediction: Genetic distance can be used to predict unobserved phenotypes which has implication in medical diagnostics, and breeding of plants and animals. *Population Genetics
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as Adaptation (biology), adaptation, s ...
: Genetic distance can help in studying population genetics, understanding intra and inter-population genetic diversity.
* Taxonomy
image:Hierarchical clustering diagram.png, 280px, Generalized scheme of taxonomy
Taxonomy is a practice and science concerned with classification or categorization. Typically, there are two parts to it: the development of an underlying scheme o ...
and Species Delimitation: Determining genetic distance through DNA barcoding
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called " sequences"), an indiv ...
is an effective tool for delimiting species especially identifying cryptic species
In biology, a species complex is a group of closely related organisms that are so similar in appearance and other features that the boundaries between them are often unclear. The taxa in the complex may be able to hybridize readily with each oth ...
. An optimized percentage threshold genetic distance is recommended based on the data and species being studied to improve and enhance the reliability and applicability of delimitation that can delineate species boundaries and identify cryptic species that look similar but are genetically distinct.
Evolutionary forces affecting genetic distance
Evolutionary forces such as mutation, genetic drift,natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generation ...
, and gene flow
In population genetics, gene flow (also known as migration and allele flow) is the transfer of genetic variation, genetic material from one population to another. If the rate of gene flow is high enough, then two populations will have equivalent ...
drive the process of evolution and genetic diversity. All these forces play significant role in genetic distance within and among species.
Measures
Different statistical measures exist that aim to quantify genetic deviation between populations or species. By utilizing assumptions gained from experimental analysis of evolutionary forces, a model that more accurately suits a given experiment can be selected to study a genetic group. Additionally, comparing how well different metrics model certain population features such as isolation can identify metrics that are more suited for understanding newly studied groups The most commonly used genetic distance metrics are Nei's genetic distance, Cavalli-Sforza and Edwards measure, and Reynolds, Weir and Cockerham's genetic distance.Jukes-Cantor Distance
One of the most basic and straight forward distance measures is Jukes-Cantor distance. This measure is constructed based on the assumption that no insertions or deletions occurred, all substitutions are independent, and that each nucleotide change is equally likely. With these presumptions, we can obtain the following equation: : where is the Jukes-Cantor distance between two sequences A, and B, and being the dissimilarity between the two sequences.Nei's standard genetic distance
In 1972, Masatoshi Nei published what came to be known as Nei's standard genetic distance. This distance has the nice property that if the rate of genetic change (amino acid substitution) is constant per year or generation then Nei's standard genetic distance (''D'') increases in proportion to divergence time. This measure assumes that genetic differences are caused bymutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
and genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
.
:
This distance can also be expressed in terms of the arithmetic mean of gene identity. Let be the probability for the two members of population having the same allele at a particular locus and be the corresponding probability in population . Also, let be the probability for a member of and a member of having the same allele. Now let , and represent the arithmetic mean
In mathematics and statistics, the arithmetic mean ( ), arithmetic average, or just the ''mean'' or ''average'' is the sum of a collection of numbers divided by the count of numbers in the collection. The collection is often a set of results fr ...
of , and over all loci, respectively. In other words,
:
:
:
where is the total number of loci examined.
Nei's standard distance can then be written as
:
Cavalli-Sforza chord distance
In 1967 Luigi Luca Cavalli-Sforza andA. W. F. Edwards
Anthony William Fairbank Edwards, Fellow of the Royal Society, FRS One or more of the preceding sentences incorporates text from the royalsociety.org website where: (born 1935) is a British statistician, geneticist and evolutionary biologist. Ed ...
published this measure. It assumes that genetic differences arise due to genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
only. One major advantage of this measure is that the populations are represented in a hypersphere, the scale of which is one unit per gene substitution. The chord distance in the hyperdimensional sphere is given by
:
Some authors drop the factor to simplify the formula at the cost of losing the property that the scale is one unit per gene substitution.
Reynolds, Weir, and Cockerham's genetic distance
In 1983, this measure was published by John Reynolds, Bruce Weir and C. Clark Cockerham. This measure assumes that genetic differentiation occurs only bygenetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
without mutations. It estimates the coancestry coefficient which provides a measure of the genetic divergence by:
:
Kimura 2 Parameter distance
 The Kimura two parameter model (K2P) was developed in 1980 by Japanese biologist Motoo Kimura. It is compatible with the neutral theory of evolution, which was also developed by the same author. As depicted in Figure 4, this measure of genetic distance accounts for the type of mutation occurring, namely whether it is a transition (i.e. purine to purine or pyrimidine to pyrimidine) or a transversion (i.e. purine to pyrimidine or vice versa). With this information, the following formula can be derived:
:
where P is and Q is , with being the number of transition type conversions, being the number of transversion type conversions, and being the number of nucleotides sites compared.
It is worth noting when transition and transversion type substitutions have an equal chance of occurring, and is assumed to equal , then the above formula can be reduced down to the Jukes Cantor model. In practice however, is typically larger than .
It has been shown that while K2P works well in classifying distantly-related species, it is not always the best choice for comparing closely-related species. In these cases, it may be better to use p-distance instead.
The Kimura two parameter model (K2P) was developed in 1980 by Japanese biologist Motoo Kimura. It is compatible with the neutral theory of evolution, which was also developed by the same author. As depicted in Figure 4, this measure of genetic distance accounts for the type of mutation occurring, namely whether it is a transition (i.e. purine to purine or pyrimidine to pyrimidine) or a transversion (i.e. purine to pyrimidine or vice versa). With this information, the following formula can be derived:
:
where P is and Q is , with being the number of transition type conversions, being the number of transversion type conversions, and being the number of nucleotides sites compared.
It is worth noting when transition and transversion type substitutions have an equal chance of occurring, and is assumed to equal , then the above formula can be reduced down to the Jukes Cantor model. In practice however, is typically larger than .
It has been shown that while K2P works well in classifying distantly-related species, it is not always the best choice for comparing closely-related species. In these cases, it may be better to use p-distance instead.
Kimura 3 Parameter distance
 The Kimura three parameter (K3P) model was first published in 1981. This measure assumes three rates of substitution when nucleotides mutate, which can be seen in Figure 5. There is one rate for transition type mutations, one rate for transversion type mutations to corresponding bases (e.g. G to C; transversion type 1 in the figure), and one rate for transversion type mutations to non-corresponding bases (e.g. G to T; transversion type 2 in the figure).
With these rates of substitution, the following formula can be derived:
:
where is the probability of a transition type mutation, is the probability of a transversion type mutation to a corresponding base, and is the probability of a transversion type mutation to a non-corresponding base. When and are assumed to be equal, this reduces down to the Kimura 2 parameter distance.
The Kimura three parameter (K3P) model was first published in 1981. This measure assumes three rates of substitution when nucleotides mutate, which can be seen in Figure 5. There is one rate for transition type mutations, one rate for transversion type mutations to corresponding bases (e.g. G to C; transversion type 1 in the figure), and one rate for transversion type mutations to non-corresponding bases (e.g. G to T; transversion type 2 in the figure).
With these rates of substitution, the following formula can be derived:
:
where is the probability of a transition type mutation, is the probability of a transversion type mutation to a corresponding base, and is the probability of a transversion type mutation to a non-corresponding base. When and are assumed to be equal, this reduces down to the Kimura 2 parameter distance.
Other measures
Many other measures of genetic distance have been proposed with varying success.Nei's ''D''''A'' distance 1983
Nei's ''D''''A'' distance was created by Masatoshi Nei, a Japanese-American biologist in 1983. This distance assumes that genetic differences arise due tomutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
and genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
, but this distance measure is known to give more reliable population trees than other distances particularly for microsatellite DNA data. This method is not ideal in cases where natural selection plays a significant role in a populations genetics.
:
: Nei's DA distance, the genetic distance between populations X and Y
: A locus or gene studied with being the sum of loci or genes
and : The frequencies of allele u in populations X and Y, respectively
L: The total number of loci examined
Euclidean distance
Euclidean distance is a formula brought about from Euclid's ''Elements, a 13 book set detailing the foundation of all euclidean mathematics. The foundational principles outlined in these works is used not only in euclidean spaces but expanded upon by Issac Newton and Gottfried Leibniz in isolated pursuits to create calculus.'' ''The euclidean distance formula is'' used to convey, as simply as possible, the genetic dissimilarity between populations, with a larger distance indicating greater dissimilarity. As seen in figure 6, this method can be visualized in a graphical manner, this is due to the work of René Descartes who created the fundamental principle of analytic geometry, or the cartesian coordinate system. In an interesting example of historical repetitions, René Descartes was not the only one who discovered the fundamental principle of analytical geometry, this principle was as discovered in an isolated pursuit by Pierre de Fermat who left his work unpublished. : : Euclidean genetic distance between populations X and Y and : Allele frequencies at locus u in populations X and Y, respectivelyGoldstein distance 1995
It was specifically developed for microsatellite markers and is based on the stepwise-mutation model (SMM). The Goldstein distance formula is modeled in such a way that expected value will increase linearly with time, this property is maintained even when the assumptions of single-step mutations and symmetrical mutation rate are violated. Goldstein distance is derived from the average square distance model, of which Goldstein was also a contributor. : :: Goldstein genetic distance between populations X and Y : and : Mean allele sizes in populations X and Y :L: Total number of microsatallite loci examinedNei's minimum genetic distance 1973
This calculation represents the minimum amount of codon differences for each locus. The measurement is based on the assumption that genetic differences arise due tomutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
and genetic drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene va ...
.
: Minimum amount of codon difference per locus
and : Average probability of two members of the X population having the same allele
: Average probability of members of the X and Y populations having the same allele
Czekanowski (Manhattan) Distance
 Similar to Euclidean distance, Czekanowski distance involves calculated the distance between points of allele frequency that are graphed on an axis created by . However, Czekanowski assumes a direct path is not available and sums the sides of the triangle formed by the data points instead of finding the hypotenuse. This formula is nicknamed the Manhattan distance because its methodology is similar to the nature of the New York City burrow. Manhattan is mainly built on a grid system requiring resentence to only make 90 degree turns during travel, which parallels the thinking of the formula.
and : Allele frequencies at locus u in populations X and Y, respectively
and : X-axis value of the frequency of an allele for X and Y populations
and : Y-axis value of the frequency of an allele for X and Y populations
Similar to Euclidean distance, Czekanowski distance involves calculated the distance between points of allele frequency that are graphed on an axis created by . However, Czekanowski assumes a direct path is not available and sums the sides of the triangle formed by the data points instead of finding the hypotenuse. This formula is nicknamed the Manhattan distance because its methodology is similar to the nature of the New York City burrow. Manhattan is mainly built on a grid system requiring resentence to only make 90 degree turns during travel, which parallels the thinking of the formula.
and : Allele frequencies at locus u in populations X and Y, respectively
and : X-axis value of the frequency of an allele for X and Y populations
and : Y-axis value of the frequency of an allele for X and Y populations
Roger's Distance 1972
Limitations of Simple Distance Formulas
While these formulas are easy and quick calculations to make, the information that is provided gives limited information. The results of these formulas do not account for the potential effects of the number of codon changes between populations, or separation time between populations.Fixation Index
A commonly used measure of genetic distance is thefixation index
The fixation index (FST) is a measure of population differentiation due to genetic structure. It is frequently estimated from Polymorphism (biology), genetic polymorphism data, such as single-nucleotide polymorphisms (SNP) or Microsatellite (genet ...
(FST) which varies between 0 and 1. A value of 0 indicates that two populations are genetically identical (minimal or no genetic diversity between the two populations) whereas a value of 1 indicates that two populations are genetically different (maximum genetic diversity between the two populations). No mutation is assumed. Large populations between which there is much migration, for example, tend to be little differentiated whereas small populations between which there is little migration tend to be greatly differentiated. FST is a convenient measure of this differentiation, and as a result FST and related statistics are among the most widely used descriptive statistics in population and evolutionary genetics. But FST is more than a descriptive statistic and measure of genetic differentiation. FST is directly related to the ''Variance'' in allele frequency among populations and conversely to the degree of resemblance among individuals within populations. If FST is small, it means that allele frequencies within each population are very similar; if it is large, it means that allele frequencies are very different.
Software
* PHYLIP useGENDIST
** Nei's standard genetic distance 1972 ** Cavalli-Sforza and Edwards 1967 ** Reynolds, Weir, and Cockerham's 1983
TFPGA
** Nei's standard genetic distance (original and unbiased) ** Nei's minimum genetic distance (original and unbiased) ** Wright's (1978) modification of Roger's (1972) distance ** Reynolds, Weir, and Cockerham's 1983
GDA
POPGENE
** Commonly used genetic distances and gene diversity analysis
** Nei's standard genetic distance 1972 ** Nei's DA distance between populations 1983
See also
* Coefficient of relationship * Degree of consanguinity * Human genetic variation *Phylogenetics
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical dat ...
* Allele frequency
Allele frequency, or gene frequency, is the relative frequency of an allele (variant of a gene) at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population tha ...
References
External links
''The Estimation of Genetic Distance and Population Substructure from Microsatellite allele frequency data.'', Brent W. Murray (May 1996), McMaster University website on genetic distance
{{DEFAULTSORT:Genetic Distance Genetics concepts Phylogenetics