DNA Base Flipping on:
[Wikipedia]
[Google]
[Amazon]
 DNA base flipping, or nucleotide flipping, is a mechanism in which a single
DNA base flipping, or nucleotide flipping, is a mechanism in which a single
 DNA can have
DNA can have
nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
base, or nucleobase
Nucleotide bases (also nucleobases, nitrogenous bases) are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nuc ...
, is rotated outside the nucleic acid double helix. This occurs when a nucleic acid-processing enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
needs access to the base to perform work on it, such as its excision for replacement with another base during DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
. It was first observed in 1994 using X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
in a methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Ro ...
enzyme catalyzing methylation of a cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
base in DNA. Since then, it has been shown to be used by different enzymes in many biological processes such as DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
, various DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
mechanisms, and DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
. It can also occur in RNA double helices or in the DNA:RNA intermediates formed during RNA transcription
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
.
DNA base flipping occurs by breaking the hydrogen bonds
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, covalently bonded to a mo ...
between the bases and unstacking the base from its neighbors. This could occur through an active process, where an enzyme binds to the DNA and then facilitates rotation of the base, or a passive process, where the
base rotates out spontaneously, and this state is recognized and bound by an enzyme. It can be detected using
X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
, NMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique based on re-orientation of atomic nuclei with non-zero nuclear spins in an external magnetic f ...
, fluorescence spectroscopy
Fluorescence spectroscopy (also known as fluorimetry or spectrofluorometry) is a type of electromagnetic spectroscopy that analyzes fluorescence from a sample. It involves using a beam of light, usually ultraviolet light, that excites the electro ...
, or hybridization probes.
Discovery
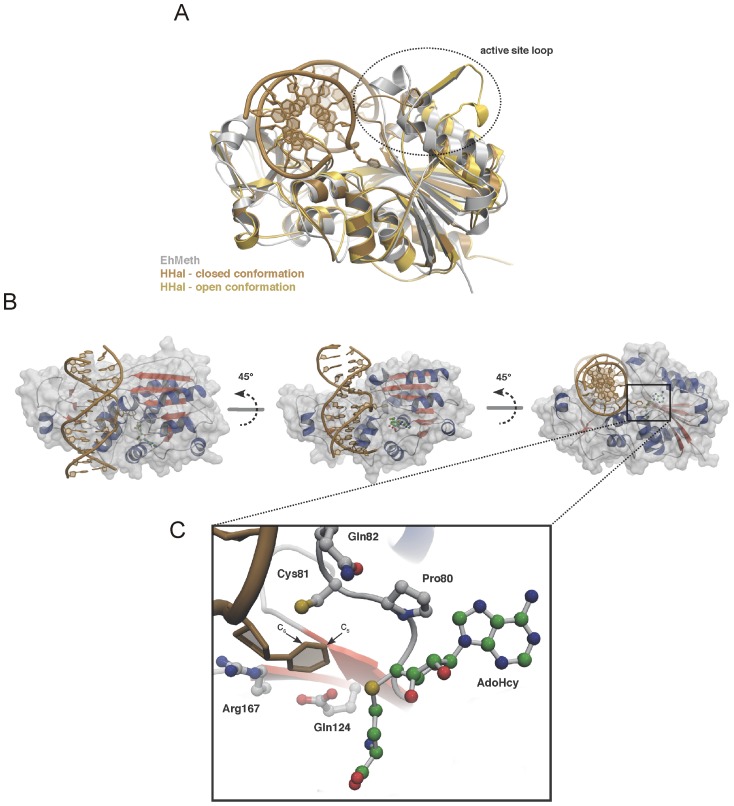
Base flipping was first observed in 1994 when researchers Klimasauskas, Kumar, Roberts, and Cheng usedX-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
to view an intermediate step in the chemical reaction of a methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Ro ...
bound to DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
. The methyltransferase they used was the C5-cytosine methyltransferase from '' Haemophilus haemolyticus'' (M. HhaI). This enzyme recognizes a specific sequence of the DNA (5'-GCGC-3') and methylates the first cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
base of the sequence at its C5 location. Upon crystallization of the M. HhaI-DNA complex, they saw the target cytosine base was rotated completely out of the double helix and was positioned in the active site of the M. HhaI. It was held in place by numerous interactions between the M. HhaI and DNA.
The authors theorized that base flipping was a mechanism used by many other enzymes, such as helicases
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized n ...
, recombination enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s, RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
s, DNA polymerases
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
, and Type II topoisomerase
Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change th ...
s. Much research has been done in the years subsequent to this discovery and it has been found that base flipping is a mechanism used in many of the biological processes the authors suggest.
Mechanism

DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
nucleotides are held together with hydrogen bonds
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, covalently bonded to a mo ...
, which are relatively weak and can be easily broken. Base flipping occurs on a millisecond timescale by breaking the hydrogen bonds between bases and unstacking the base from its neighbors. The base is rotated out of the double helix
In molecular biology, the term double helix refers to the structure formed by base pair, double-stranded molecules of nucleic acids such as DNA. The double Helix, helical structure of a nucleic acid complex arises as a consequence of its Nuclei ...
by 180 degrees, typically via the major groove
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, a ...
, and into the active site of an enzyme. This opening leads to small conformational changes in the DNA backbone which are quickly stabilized by the increased enzyme-DNA interactions. Studies looking at the free-energy profiles of base flipping have shown that the free-energy barrier to flipping can be lowered by 17 kcal/mol for M.HhaI in the closed conformation.
There are two mechanisms of DNA base flipping: active and passive. In the active mechanism, an enzyme binds to the DNA and then actively rotates the base, while in the passive mechanism a damaged base rotates out spontaneously first, then is recognized and bound by the enzyme. Research has demonstrated both mechanisms: uracil-DNA glycosylase
Uracil-DNA glycosylase (also known as UNG or UDG) is an enzyme. Its most important function is to prevent mutagenesis by eliminating uracil from DNA molecules by cleaving the N-glycosidic bond and initiating the base-excision repair (BER) pathway ...
follows the passive mechanism and Tn10 Tn''10'' is a transposable element, which is a sequence of DNA that is capable of mediating its own movement from one position in the DNA of the host organism to another. There are a number of different transposition mechanisms in nature, but Tn'' ...
transposase
A transposase is any of a class of enzymes capable of binding to the end of a transposon and catalysing its movement to another part of a genome, typically by a cut-and-paste mechanism or a replicative mechanism, in a process known as transpositio ...
follows the active mechanism.
Furthermore, studies have shown that DNA base flipping is used by many different enzymes in a variety biological processes such as DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
, various DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
mechanisms, RNA transcription
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
and DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
.
Biological processes
DNA modification and repair
 DNA can have
DNA can have mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s that cause a base in the DNA strand to be damaged. To ensure genetic integrity of the DNA, enzymes need to repair any damage. There are many types of DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
. Base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from t ...
utilizes base flipping to flip the damaged base out of the double helix and into the specificity pocket of a glycosylase Glycosylases ( EC 3.2) are enzymes that hydrolyze glycosyl compounds. They are a type of hydrolase (EC 3). In turn, glycosylases are divided into two groups: glycosidase
In biochemistry, glycoside hydrolases (also called glycosidases or glyco ...
which hydrolyzes the glycosidic bond and removes the base. DNA glycosylases interact with DNA, flipping bases to determine a mismatch. An example of base excision repair occurs when a cytosine base is deaminated and becomes a uracil base. This causes a U:G mispair which is detected by Uracil DNA glycosylase. The uracil base is flipped out into the glycosylase active pocket where it is removed from the DNA strand. Base flipping is used to repair mutations such as 8-Oxoguanine (oxoG) and thymine dimers created by UV radiation.
Replication, transcription and recombination
DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
and RNA transcription
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
both make use of base flipping. DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
is an enzyme that carries out replication. It can be thought of as a hand that grips the DNA single strand template. As the template passes across the palm region of the polymerase, the template bases are flipped out of the helix and away from the dNTP
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chai ...
binding site. During transcription, RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
catalyzes RNA synthesis. During the initiation
Initiation is a rite of passage marking entrance or acceptance into a group or society. It could also be a formal admission to adulthood in a community or one of its formal components. In an extended sense, it can also signify a transformatio ...
phase, two bases in the -10 element flip out from the helix and into two pockets in RNA polymerase. These new interactions stabilize the -10 element and promote the DNA strands to separate or melt.
Base flipping occurs during latter stages of recombination. RecA
RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA in bacteria. Structural and functional homologs to RecA have been found in all kingdoms of life. RecA serves as an archetype for this class of homologous DNA repair p ...
is a protein that promotes strand invasion during homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organi ...
. Base flipping has been proposed as the mechanism by which RecA can enable a single strand to recognize homology in duplex DNA. Other studies indicate that it is also involved in V(D)J Recombination
V(D)J recombination (variable–diversity–joining rearrangement) is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire ...
.
DNA methylation

DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
is the process in which a methyl group
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula (whereas normal methane has the formula ). In formulas, the group is often abbreviated a ...
is added to either a cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
or adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
. This process causes the activation or inactivation of gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
, thereby resulting in gene regulation in eukaryotic cells. DNA methylation process is also known to be involved in certain types of cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
formation. In order for this chemical modification to occur, it is necessary that the target base flips out of the DNA double helix to allow the methyltransferases to catalyze the reaction.
Target recognition by restriction endonucleases
Restriction endonucleases, also known asrestriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
s are enzymes that cleave the sugar-phosphate backbone of the DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
at specific nucleotides
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
sequences that are usually four to six nucleotides long. Studies performed by Horton and colleagues have shown that the mechanism by which these enzymes cleave the DNA involves base flipping as well as bending the DNA and the expansion of the minor groove
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, a ...
. In 2006, Horton and colleagues, x-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
evidence was presented showing that the restriction endonuclease HinP1I utilizes base flipping in order to recognize its target sequence. This enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
is known to cleave the DNA at the palindromic
A palindrome ( /ˈpæl.ɪn.droʊm/) is a word, number, phrase, or other sequence of symbols that reads the same backwards as forwards, such as ''madam'' or '' racecar'', the date " 02/02/2020" and the sentence: "A man, a plan, a canal – Pana ...
tetranucleotide sequence G↓CGC.
Experimental approaches for detection
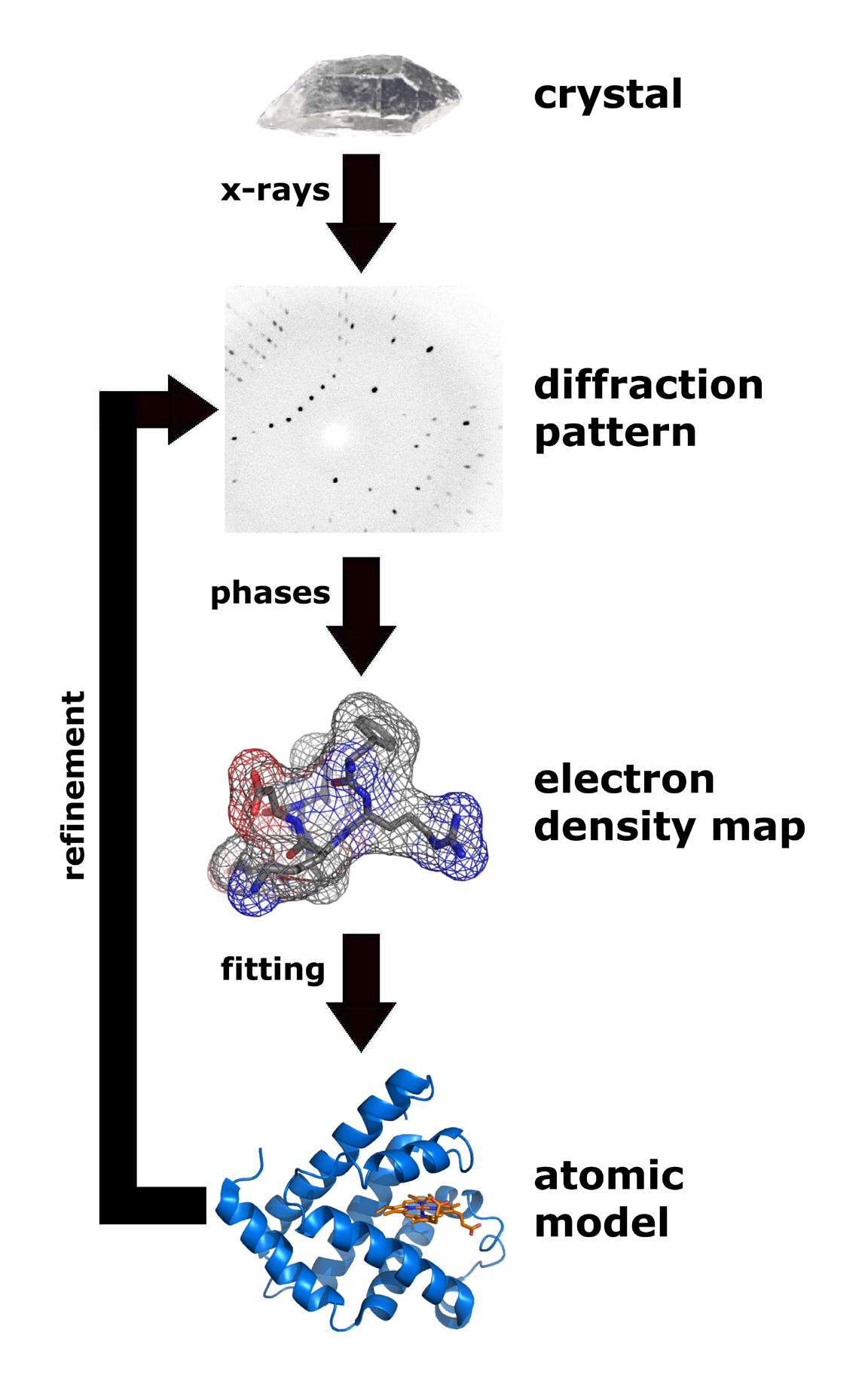
X-ray crystallography

X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring th ...
is a technique that measures the angles and intensities of crystalline atoms in order to determine the atomic and molecular structure of the crystal of interest. Crystallographers are then able to produce and three-dimensional picture where the positions of the atoms
Atoms are the basic particles of the chemical elements. An atom consists of a nucleus of protons and generally neutrons, surrounded by an electromagnetically bound swarm of electrons. The chemical elements are distinguished from each other ...
, chemical bonds
A chemical bond is the association of atoms or ions to form molecules, crystals, and other structures. The bond may result from the electrostatic force between oppositely charged ions as in ionic bonds or through the sharing of electrons as ...
as well as other important characteristics can be determined. Klimasaukas and colleagues used this technique to observe the first base flipping phenomenon, in which their experimental procedure involved several steps:
# Purification
# Crystallization
# Data Collection
# Structure determination and refinement
During purification, Haemophilus haemolyticus methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Ro ...
was overexpressed and purified using a high salt back-extraction step to selectively solubilize M.HhaI, followed by fast protein liquid chromatography ( FPLC) as done previously by Kumar and colleagues. Authors utilized a Mono-Q anion exchange column to remove the small quantity of proteinaceous
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respon ...
materials and unwanted DNA prior to the crystallization step. Once M.HhaI was successfully purified, the sample was then grown using a method that mixes the solution containing the complex at a temperature of 16 °C and the hanging-drop vapor diffusion technique to obtain the crystals. Authors were then able to collect the x-ray data according to a technique used by Cheng and colleagues in 1993. This technique involved the measurement of the diffraction intensities on a FAST detector, where the exposure times for 0.1° rotation were 5 or 10 seconds. For the structure determination and refinement, Klimasaukas and colleagues used the molecular replacement of the refined apo structure described by Cheng and colleagues in 1993 where the search models X-PLOR
X-PLOR is a computer software package for computational structural biology originally developed by Axel T. Brunger at Yale University. It was first published in 1987 as an offshoot of CHARMM - a similar program that ran on supercomputers made by C ...
, MERLOT
Merlot ( ) is a dark-blue-colored wine grape variety that is used as both a blending grape and for varietal wines. The name ''Merlot'' is thought to be a diminutive of , the French name for the blackbird, probably a reference to the color ...
, and TRNSUM were used to solve the rotation and translation functions. This part of the study involves the use of a variety of software and computer algorithms to solve the structures and characteristics of the crystal of interest.
NMR spectroscopy
NMR spectroscopy
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique based on re-orientation of atomic nuclei with non-zero nuclear spins in an external magnetic f ...
is a technique that has been used over the years to study important dynamic aspects of base flipping. This technique allows researchers to determine the physical and chemical properties of atoms
Atoms are the basic particles of the chemical elements. An atom consists of a nucleus of protons and generally neutrons, surrounded by an electromagnetically bound swarm of electrons. The chemical elements are distinguished from each other ...
and other molecules by utilizing the magnetic properties of atomic nuclei
The atomic nucleus is the small, dense region consisting of protons and neutrons at the center of an atom, discovered in 1911 by Ernest Rutherford at the University of Manchester based on the 1909 Geiger–Marsden gold foil experiment. Aft ...
. In addition, NMR can provide a variety of information including structure, reaction states, chemical environment of the molecules, and dynamics. During the DNA base flipping discovery experiment, researchers utilized NMR spectroscopy to investigate the enzyme-induced base flipping of HhaI methyltransferase. In order to accomplish this experiment, two 5-fluorocytosine residues were incorporated into the target and the reference position with the DNA substrate so the 19F chemical shift analysis could be performed. Once the 19F chemical shift analysis was evaluated, it was then concluded that the DNA complexes existed with multiple forms of the target 5-fluorocytosine along the base flipping pathway.
Fluorescence spectroscopy
Fluorescence spectroscopy
Fluorescence spectroscopy (also known as fluorimetry or spectrofluorometry) is a type of electromagnetic spectroscopy that analyzes fluorescence from a sample. It involves using a beam of light, usually ultraviolet light, that excites the electro ...
is a technique that is used to assay a sample using a fluorescent probe. DNA nucleotides themselves are not good candidates for this technique because they do not readily re-emit light upon light excitation. A fluorescent marker is needed to detect base flipping. 2-Aminopurine
2-Aminopurine (2AP), a Base analog, purine analog of guanine and adenine, is a fluorescent molecular marker used in nucleic acid research. It most commonly pairs with thymine as an adenine-analogue. It uses a different ketone oxygen on thymine fo ...
is a base that is structurally similar to adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
, but is very fluorescent when flipped out from the DNA duplex. It is commonly used to detect base flipping and has an excitation at 305‑320 nm and emission at 370 nm so that it well separated from the excitations of proteins and DNA. Other fluorescent probes used to study DNA base flipping are 6MAP (4‑amino‑6‑methyl‑7(8H)‑pteridone) and Pyrrolo‑C (3- �-D-2-ribofuranosyl6-methylpyrrolo ,3-dyrimidin-2(3H)-one). Time-resolved fluorescence spectroscopy is also employed to provide a more detailed picture of the extent of base flipping as well as the conformational dynamics occurring during base flipping.
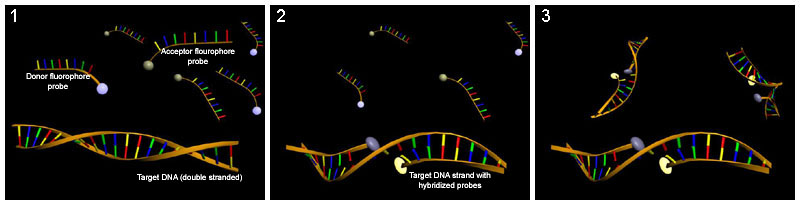
Hybridization probing
Hybridization probes can be used to detect base flipping. This technique uses a molecule that has a complementary sequence to the sequence you would like to detect such that it binds to a single-strand of the DNA or RNA. Several hybridization probes have been used to detect base flipping.Potassium permanganate
Potassium permanganate is an inorganic compound with the chemical formula KMnO4. It is a purplish-black crystalline salt, which dissolves in water as K+ and ions to give an intensely pink to purple solution.
Potassium permanganate is widely us ...
is used to detect thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
residues that have been flipped out by cytosine-C5 and adenine-N6 methyltransferases. Chloroacetaldehyde
Chloroacetaldehyde is an organic compound with the formula ClCH2CHO. Like some related compounds, it is highly electrophilic reagent and a potentially dangerous alkylating agent. The compound is not normally encountered in the anhydrous form, but ...
is used to detect cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
residues flipped out by the HhaI DNA cytosine-5 methyltransferase (M. HhaI).

See also
*DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
* Base excision repair
Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from t ...
* DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
* RNA transcription
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
* DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter (genetics), promoter, DNA methylati ...
* DNA methyltransferase
In biochemistry, the DNA methyltransferase (DNA MTase, DNMT) family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl ...
* Genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryot ...
* Homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organi ...
* DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
* Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
* Epigenomics
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome. The field is analogous to genomics and proteomics, which are the study of the genome and proteome of a cell. Epige ...
References
{{reflist, 2 Molecular biology Base flipping