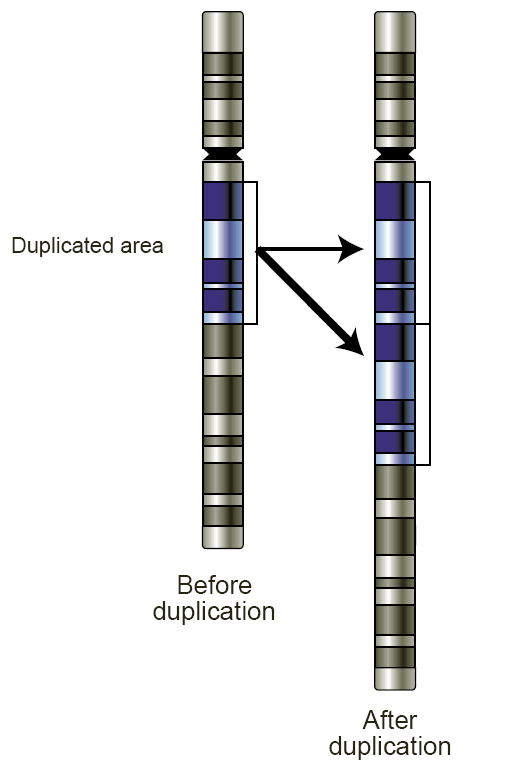
 Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammal
A mammal () is a vertebrate animal of the Class (biology), class Mammalia (). Mammals are characterised by the presence of milk-producing mammary glands for feeding their young, a broad neocortex region of the brain, fur or hair, and three ...
s, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype.
Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Long repeats include repeats of entire genes. This classification based on size of the repeat is the most obvious type of classification as size is an important factor in examining the types of mechanisms that most likely gave rise to the repeats, hence the likely effects of these repeats on phenotype.
Types and chromosomal rearrangements
One of the most well known examples of a short copy number variation is the trinucleotide repeat of the CAG base pairs in the huntingtin gene responsible for the neurological disorderHuntington's disease
Huntington's disease (HD), also known as Huntington's chorea, is an incurable neurodegenerative disease that is mostly Genetic disorder#Autosomal dominant, inherited. It typically presents as a triad of progressive psychiatric, cognitive, and ...
. For this particular case, once the CAG trinucleotide repeats more than 36 times in a trinucleotide repeat expansion, Huntington's disease will likely develop in the individual and it will likely be inherited by his or her offspring. The number of repeats of the CAG trinucleotide is inversely correlated with the age of onset of Huntington's disease. These types of short repeats are often thought to be due to errors in polymerase
In biochemistry, a polymerase is an enzyme (Enzyme Commission number, EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by ...
activity during replication including polymerase slippage, template switching, and fork switching which will be discussed in detail later. The short repeat size of these copy number variations lends itself to errors in the polymerase as these repeated regions are prone to misrecognition by the polymerase and replicated regions may be replicated again, leading to extra copies of the repeat. In addition, if these trinucleotide repeats are in the same reading frame in the coding portion of a gene, it may lead to a long chain of the same amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
, possibly creating protein aggregates in the cell, and if these short repeats fall into the non-coding portion of the gene, it may affect gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
and regulation. On the other hand, a variable number of repeats of entire genes is less commonly identified in the genome. One example of a whole gene repeat is the alpha-amylase 1 gene (''AMY1'') that encodes alpha-amylase which has a significant copy number variation between different populations with different diets. Although the specific mechanism that allows the ''AMY1'' gene to increase or decrease its copy number is still a topic of debate, some hypotheses suggest that the non-homologous end joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. It is called "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair ...
or the microhomology-mediated end joining is likely responsible for these whole gene repeats. Repeats of entire genes has immediate effects on expression of that particular gene, and the fact that the copy number variation of the ''AMY1'' gene has been related to diet is a remarkable example of recent human evolutionary adaptation. Although these are the general groups that copy number variations are grouped into, the exact number of base pairs copy number variations affect depends on the specific loci of interest. Currently, using data from all reported copy number variations, the mean size of copy number variant is around 118kb, and the median is around 18kb.
In terms of the structural architecture of copy number variations, research has suggested and defined hotspot regions in the genome where copy number variations are four times more enriched. These hotspot regions were defined to be regions containing long repeats that are 90–100% similar known as segmental duplications either tandem
Tandem, or in tandem, is an arrangement in which two or more animals, machines, or people are lined up one behind another, all facing in the same direction. ''Tandem'' can also be used more generally to refer to any group of persons or objects w ...
or interspersed and most importantly, these hotspot regions have an increased rate of chromosomal rearrangement. It was thought that these large-scale chromosomal rearrangements give rise to normal variation and genetic diseases, including copy number variations. Moreover, these copy number variation hotspots are consistent throughout many populations from different continents, implying that these hotspots were either independently acquired by all the populations and passed on through generations, or they were acquired in early human evolution before the populations split, the latter seems more likely. Lastly, spatial biases of the location at which copy number variations are most densely distributed does not seem to occur in the genome. Although it was originally detected by fluorescent in situ hybridization and microsatellite analysis that copy number repeats are localized to regions that are highly repetitive such as telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In ...
s, centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fiber ...
s, and heterochromatin, recent genome-wide studies have concluded otherwise. Namely, the subtelomeric regions and pericentromeric regions are where most chromosomal rearrangement hotspots are found, and there is no considerable increase in copy number variations in that region. Furthermore, these regions of chromosomal rearrangement hotspots do not have decreased gene numbers, again, implying that there is minimal spatial bias of the genomic location of copy number variations.
Detection and identification
Copy number variation was initially thought to occupy an extremely small and negligible portion of the genome through cytogenetic observations. Copy number variations were generally associated only with small tandem repeats or specific genetic disorders, therefore, copy number variations were initially only examined in terms of specific loci. However, technological developments led to an increasing number of highly accurate ways of identifying and studying copy number variations. Copy number variations were originally studied by cytogenetic techniques, which are techniques that allow one to observe the physical structure of the chromosome. One of these techniques is fluorescent in situ hybridization (FISH) which involves inserting fluorescent probes that require a high degree of complementarity in the genome for binding. Comparative genomic hybridization was also commonly used to detect copy number variations byfluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with se ...
visualization and then comparing the length of the chromosomes.
Recent advances in genomics
Genomics is an interdisciplinary field of molecular biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, ...
technologies gave rise to many important methods that are of extremely high genomic resolution and as a result, an increasing number of copy number variations in the genome have been reported. Initially these advances involved using bacterial artificial chromosome
A bacterial artificial chromosome (BAC) is a DNA construct, based on a functional fertility plasmid (or F-plasmid), used for transforming and cloning in bacteria, usually '' E. coli''. F-plasmids play a crucial role because they contain partiti ...
(BAC) array with around 1 megabase of intervals throughout the entire gene, BACs can also detect copy number variations in rearrangement hotspots allowing for the detection of 119 novel copy number variations. High throughput genomic sequencing has revolutionized the field of human genomics and in silico
In biology and other experimental sciences, an ''in silico'' experiment is one performed on a computer or via computer simulation software. The phrase is pseudo-Latin for 'in silicon' (correct ), referring to silicon in computer chips. It was c ...
studies have been performed to detect copy number variations in the genome. Reference sequences have been compared to other sequences of interest using fosmids by strictly controlling the fosmid clones to be 40kb. Sequencing end reads would provide adequate information to align the reference sequence to the sequence of interest, and any misalignments are easily noticeable thus concluded to be copy number variations within that region of the clone. This type of detection technique offers a high genomic resolution and precise location of the repeat in the genome, and it can also detect other types of structural variation such as inversions.
In addition, another way of detecting copy number variation is using single nucleotide polymorphisms
In genetics and bioinformatics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in ...
(SNPs). Due to the abundance of the human SNP data, the direction of detecting copy number variation has changed to utilize these SNPs. Relying on the fact that human recombination is relatively rare and that many recombination events occur in specific regions of the genome known as recombination hotspots, linkage disequilibrium can be used to identify copy number variations. Efforts have been made in associating copy number variations with specific haplotype
A haplotype (haploid genotype) is a group of alleles in an organism that are inherited together from a single parent.
Many organisms contain genetic material (DNA) which is inherited from two parents. Normally these organisms have their DNA orga ...
SNPs by analyzing the linkage disequilibrium, using these associations, one is able to recognize copy number variations in the genome using SNPs as markers. Next-generation sequencing techniques including short and long read sequencing are nowadays increasingly used and have begun to replace array-based techniques to detect copy number variations.
Molecular mechanism
There are two main types of molecular mechanism for the formation of copy number variations: homologous based and non-homologous based. Although many suggestions have been put forward, most of these theories are speculations and conjecture. There is no conclusive evidence that correlates a specific copy number variation to a specific mechanism.cohesin
Cohesin is a protein complex that mediates Establishment of sister chromatid cohesion, sister chromatid cohesion, homologous recombination, and Topologically associating domain, DNA looping. Cohesin is formed of SMC3, SMC1A, SMC1, RAD21, SCC1 an ...
proteins are found to aid in the repair system of double stranded breaks through clamping the two ends in close proximity which prevents interchromosomal invasion of the ends. If for any reason, such as activation of ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal ...
, cohesin activity is affected then there may be local increase in double stranded break repair errors.
The other class of possible mechanisms that are hypothesized to lead to copy number variations is non-homologous based. To distinguish between this and homologous based mechanisms, one must understand the concept of homology. Homologous pairing of chromosomes involved using DNA strands that are highly similar to each other (~97%) and these strands must be longer than a certain length to avoid short but highly similar pairings. Non-homologous pairings, on the other hand, rely on only few base pairs of similarity between two strands, therefore it is possible for genetic materials to be exchanged or duplicated in the process of non-homologous based double stranded repairs.
One type of non-homologous based mechanism is the non-homologous end joining or micro-homology end joining mechanism. These mechanisms are also involved in repairing double stranded breaks but require no homology or limited micro-homology. When these strands are repaired, oftentimes there are small deletions or insertions added into the repaired strand. It is possible that retrotransposons
Retrotransposons (also called Class I transposable elements) are transposable element, mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II trans ...
are inserted into the genome through this repair system. If retrotransposons are inserted into a non-allelic position on the chromosome, meiotic recombination can drive the insertion to be recombined into the same strand as an already existing copy of the same region. Another mechanism is the break-fusion-bridge cycle which involves sister chromatids
A sister chromatid refers to the identical copies ( chromatids) formed by the DNA replication of a chromosome, with both copies joined together by a common centromere. In other words, a sister chromatid may also be said to be 'one-half' of the du ...
that have both lost its telomeric region due to double stranded breaks. It is proposed that these sister chromatids will fuse together to form one dicentric chromosome, and then segregate into two different nuclei. Because pulling the dicentric chromosome apart causes a double stranded break, the end regions can fuse to other double stranded breaks and repeat the cycle. The fusion of two sister chromatids can cause inverted duplication and when these events are repeated throughout the cycle, the inverted region will be repeated leading to an increase in copy number. The last mechanism that can lead to copy number variations is polymerase slippage, which is also known as template switching. During normal DNA replication, the polymerase on the lagging strand is required to unclamp and re-clamp the replication region continuously. When small scale repeats in the DNA sequence exist already, the polymerase can be 'confused' when it re-clamps to continue replication and instead of clamping to the correct base pairs, it may shift a few base pairs and replicate a portion of the repeated region again. Note that although this has been experimentally observed and is a widely accepted mechanism, the molecular interactions that led to this error remains unknown. In addition, because this type of mechanism requires the polymerase to jump around the DNA strand and it is unlikely that the polymerase can re-clamp at another locus some kilobases apart, therefore this is more applicable to short repeats such as dinucleotide or trinucleotide repeats.
Alpha-amylase gene
 Amylase is an
Amylase is an enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
in saliva that is responsible for the breakdown of starch
Starch or amylum is a polymeric carbohydrate consisting of numerous glucose units joined by glycosidic bonds. This polysaccharide is produced by most green plants for energy storage. Worldwide, it is the most common carbohydrate in human diet ...
into monosaccharides, and one type of amylase is encoded by the alpha-amylase gene (''AMY1''). The ''AMY1'' locus, as well as the amylase enzyme, is one of the most extensively studied and sequenced genes in the human genome. Its homologs are also found in other primates and therefore it is likely that the primate
Primates is an order (biology), order of mammals, which is further divided into the Strepsirrhini, strepsirrhines, which include lemurs, galagos, and Lorisidae, lorisids; and the Haplorhini, haplorhines, which include Tarsiiformes, tarsiers a ...
''AMY1'' gene is ancestral to the human ''AMY1'' gene and was adapted early in primate evolution. ''AMY1'' is one of the most well studied genes which has wide range of variable numbers of copies throughout different human populations. The ''AMY1'' gene is also one of the few genes that had been studied that displayed convincing evidence which correlates its protein function to its copy number. Copy number is known to alter transcription as well as translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
levels of a particular gene, however research has shown that the relationship between protein levels and copy number is variable. In the ''AMY1'' genes of European Americans it is found that the concentration of salivary amylase is closely correlated to the copy number of the ''AMY1'' gene. As a result, it was hypothesized that the copy number of the ''AMY1'' gene is closely correlated with its protein function, which is to digest starch.
The ''AMY1'' gene copy number has been found to be correlated to different levels of starch in diets of different populations. Eight populations from different continents were categorized into high starch diets and low starch diets and their ''AMY1'' gene copy number was visualized using high resolution FISH and qPCR. It was found that the high starch diet populations which consists of the Japanese, Hadza, and European American populations had a significantly higher (two times higher) average ''AMY1'' copy number than the low starch diet populations including Biaka, Mbuti, Datog and Yakut populations. It was hypothesized that the levels of starch in one’s regular diet, the substrate for AMY1, can directly affect the copy number of the ''AMY1'' gene. Since it was concluded that the copy number of ''AMY1'' is directly correlated with salivary amylase, the more starch present in the population’s daily diet, the more evolutionarily favorable it is to have multiple copies of the ''AMY1'' gene. The ''AMY1'' gene was the first gene to provide strong evidence for evolution on a molecular genetic level. Moreover, using comparative genomic hybridization, copy number variations of the entire genomes of the Japanese population was compared to that of the Yakut population. It was found that the copy number variation of the ''AMY1'' gene was significantly different from the copy number variation in other genes or regions of the genome, suggesting that the ''AMY1'' gene was under a strong selective pressure that had little or no influence on the other copy number variations. Finally, the variability of length of 783 microsatellite
A microsatellite is a tract of repetitive DNA in which certain Sequence motif, DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organ ...
s between the two populations was compared to copy number variability of the ''AMY1'' gene. It was found that the ''AMY1'' gene copy number range was larger than that of over 97% of the microsatellites examined. This implies that natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generation ...
played a considerable role in shaping the average number of ''AMY1'' genes in these two populations. However, as only six populations were studied, it is important to consider the possibility that there may be other factors in their diet or culture that influenced the ''AMY1'' copy number other than starch.
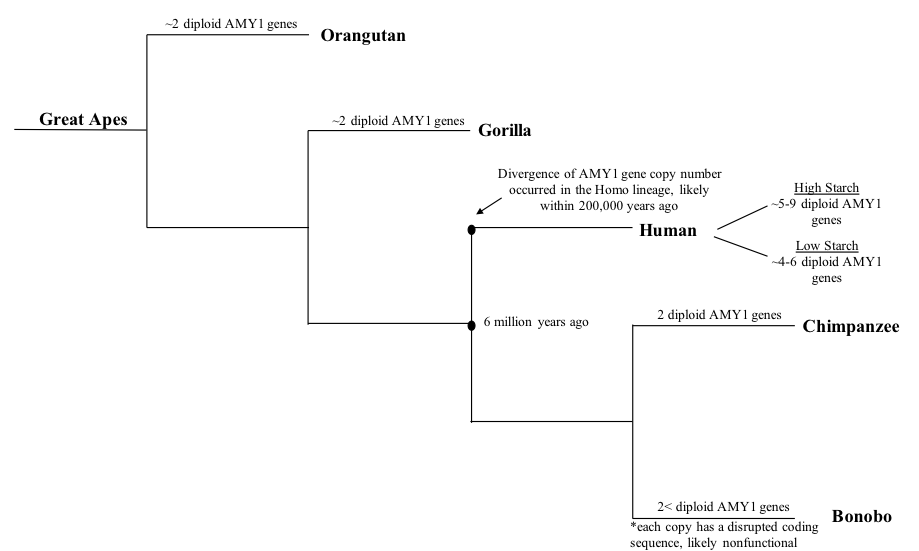 Although it is unclear when the ''AMY1'' gene copy number began to increase, it is known and confirmed that the ''AMY1'' gene existed in early primates.
Although it is unclear when the ''AMY1'' gene copy number began to increase, it is known and confirmed that the ''AMY1'' gene existed in early primates. Chimpanzee
The chimpanzee (; ''Pan troglodytes''), also simply known as the chimp, is a species of Hominidae, great ape native to the forests and savannahs of tropical Africa. It has four confirmed subspecies and a fifth proposed one. When its close rel ...
s, the closest evolutionary relatives to humans, were found to have two diploid
Ploidy () is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Here ''sets of chromosomes'' refers to the number of maternal and paternal chromosome copies, ...
copies of the ''AMY1'' gene that is identical in length to the human AMY1 gene, which is significantly less than that of humans. On the other hand, bonobo
The bonobo (; ''Pan paniscus''), also historically called the pygmy chimpanzee (less often the dwarf chimpanzee or gracile chimpanzee), is an endangered great ape and one of the two species making up the genus ''Pan (genus), Pan'' (the other bei ...
s, also a close relative of modern humans, were found to have more than two diploid copies of the ''AMY1'' gene. Nonetheless, the bonobo ''AMY1'' genes were sequenced and analyzed, and it was found that the coding sequences of the ''AMY1'' genes were disrupted, which may lead to the production of dysfunctional salivary amylase. It can be inferred from the results that the increase in bonobo ''AMY1'' copy number is likely not correlated to the amount of starch in their diet. It was further hypothesized that the increase in copy number began recently during early hominin
The Hominini (hominins) form a taxonomic tribe of the subfamily Homininae (hominines). They comprise two extant genera: ''Homo'' (humans) and '' Pan'' (chimpanzees and bonobos), and in standard usage exclude the genus '' Gorilla'' ( gorillas) ...
evolution as none of the great apes had more than two copies of the ''AMY1'' gene that produced functional protein. In addition, it was speculated that the increase in the ''AMY1'' copy number began around 20,000 years ago when humans shifted from a hunter-gatherer
A hunter-gatherer or forager is a human living in a community, or according to an ancestrally derived Lifestyle, lifestyle, in which most or all food is obtained by foraging, that is, by gathering food from local naturally occurring sources, esp ...
lifestyle to agricultural
Agriculture encompasses crop and livestock production, aquaculture, and forestry for food and non-food products. Agriculture was a key factor in the rise of sedentary human civilization, whereby farming of domesticated species created f ...
societies, which was also when humans relied heavily on root vegetables
Root vegetables are underground plant parts eaten by humans or animals as food. In agricultural and culinary terminology, the term applies to true roots, such as taproots and tuberous root, root tubers, as well as non-roots such as bulbs, corms, ...
high in starch. This hypothesis, although logical, lacks experimental evidence due to the difficulties in gathering information on the shift of human diets, especially on root vegetables that are high in starch as they cannot be directly observed or tested. Recent breakthroughs in DNA sequencing has allowed researchers to sequence older DNA such as that of Neanderthal
Neanderthals ( ; ''Homo neanderthalensis'' or sometimes ''H. sapiens neanderthalensis'') are an extinction, extinct group of archaic humans who inhabited Europe and Western and Central Asia during the Middle Pleistocene, Middle to Late Plei ...
s to a certain degree of accuracy. Perhaps sequencing Neanderthal DNA can provide a time marker as to when the ''AMY1'' gene copy number increased and offer insight into human diet and gene evolution.
Currently it is unknown which mechanism gave rise to the initial duplication of the amylase gene, and it can imply that the insertion of the retroviral sequences was due to non-homologous end joining, which caused the duplication of the ''AMY1'' gene. However, there is currently no evidence to support this theory and therefore this hypothesis remains conjecture. The recent origin of the multi-copy ''AMY1'' gene implies that depending on the environment, the ''AMY1'' gene copy number can increase and decrease very rapidly relative to genes that do not interact as directly with the environment. The ''AMY1'' gene is an excellent example of how gene dosage affects the survival of an organism in a given environment. The multiple copies of the ''AMY1'' gene give those who rely more heavily on high starch diets an evolutionary advantage, therefore the high gene copy number persists in the population.
Brain cells
Among theneuron
A neuron (American English), neurone (British English), or nerve cell, is an membrane potential#Cell excitability, excitable cell (biology), cell that fires electric signals called action potentials across a neural network (biology), neural net ...
s in the human brain
The human brain is the central organ (anatomy), organ of the nervous system, and with the spinal cord, comprises the central nervous system. It consists of the cerebrum, the brainstem and the cerebellum. The brain controls most of the activi ...
, somatically derived copy number variations are frequent. Copy number variations show wide variability (9 to 100% of brain neurons in different studies). Most alterations are between 2 and 10 Mb in size with deletions far outnumbering amplifications.
Genomic duplication and triplication of the gene appear to be a rare cause of Parkinson's disease
Parkinson's disease (PD), or simply Parkinson's, is a neurodegenerative disease primarily of the central nervous system, affecting both motor system, motor and non-motor systems. Symptoms typically develop gradually and non-motor issues become ...
, although more common than point mutations.
Copy number variants in ''RCL1
RNA 3'-terminal phosphate cyclase-like protein is an enzyme that in humans is encoded by the ''RCL1'' gene.
Copy number variants to the RCL1 gene are associated with a range of neuropsychiatric phenotypes, and a missense variant associated with ...
'' gene are associated with a range of neuropsychiatric phenotypes in children.
Gene families and natural selection
 Recently, there had been discussion connecting copy number variations to
Recently, there had been discussion connecting copy number variations to gene families
A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions. One such family are the genes for human hemoglobin subunits; the ten genes are in two clusters on ...
. Gene families are defined as a set of related genes that serve similar functions but have minor temporal or spatial differences and these genes likely derived from one ancestral gene. The main reason copy number variations are connected to gene families is that there is a possibility that genes in a family may have derived from one ancestral gene which got duplicated into different copies. Mutations accumulate through time in the genes and with natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generation ...
acting on the genes, some mutations lead to environmental advantages allowing those genes to be inherited and eventually clear gene families are separated out. An example of a gene family that may have been created due to copy number variations is the globin
The globins are a superfamily of heme-containing globular proteins, involved in binding and/or transporting oxygen. These proteins all incorporate the globin fold, a series of eight alpha helical segments. Two prominent members include myo ...
gene family. The globin gene family is an elaborate network of genes consisting of alpha
Alpha (uppercase , lowercase ) is the first letter of the Greek alphabet. In the system of Greek numerals, it has a value of one. Alpha is derived from the Phoenician letter ''aleph'' , whose name comes from the West Semitic word for ' ...
and beta
Beta (, ; uppercase , lowercase , or cursive ; or ) is the second letter of the Greek alphabet. In the system of Greek numerals, it has a value of 2. In Ancient Greek, beta represented the voiced bilabial plosive . In Modern Greek, it represe ...
globin genes including genes that are expressed in both embryos and adults as well as pseudogenes. These globin genes in the globin family are all well conserved and only differ by a small portion of the gene, indicating that they were derived from a common ancestral gene, perhaps due to duplication of the initial globin gene.
Research has shown that copy number variations are significantly more common in genes that encode proteins that directly interact with the environment than proteins that are involved in basic cellular activities. It was suggested that the gene dosage effect accompanying copy number variation may lead to detrimental effects if essential cellular functions are disrupted, therefore proteins involved in cellular pathways are subjected to strong purifying selection. In addition, proteins function together and interact with proteins of other pathways, therefore it is important to view the effects of natural selection on bio-molecular pathways rather than on individual proteins. With that being said, it was found that proteins in the periphery of the pathway are enriched in copy number variations whereas proteins in the center of the pathways are depleted in copy number variations. It was explained that proteins in the periphery of the pathway interact with fewer proteins and so a change in protein dosage affected by a change in copy number may have a smaller effect on the overall outcome of the cellular pathway.
See also
* CaSNP, a database *Comparative genomics
Comparative genomics is a branch of biological research that examines genome sequences across a spectrum of species, spanning from humans and mice to a diverse array of organisms from bacteria to chimpanzees. This large-scale holistic approach c ...
* Copy number analysis
Copy number analysis is the process of analyzing data produced by a test for DNA copy number variation in an organism's sample. One application of such analysis is the detection of chromosomal copy number variation that may cause or may increase ...
* Human genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as the DNA within each of the 23 distinct chromosomes in the cell nucleus. A small DNA molecule is found within individual Mitochondrial DNA, mitochondria. These ar ...
* Inparanoid
* Molecular evolution
Molecular evolution describes how Heredity, inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of Cell (biology), cells and organisms. Molecular evolution is the basis of phylogen ...
* Pseudogenes
* Segmental duplication
* Tandem exon duplication
* Virtual karyotype
References
Further reading
* * * * * * * * *External links
Copy Number Variation Project
Sanger Institute
The Wellcome Sanger Institute, previously known as The Sanger Centre and Wellcome Trust Sanger Institute, is a non-profit organisation, non-profit British genomics and genetics research institute, primarily funded by the Wellcome Trust.
It is l ...
The Claim: Identical Twins Have Identical DNA
Integrative annotation platform for copy number variations in humans
A bibliography on copy number variation
Database of Genomic Variants
a database of structural variants in the human genome
Copy Number Variation Detection via High-Density SNP Genotyping
BioDiscovery Nexus Copy Number
High-resolution mapping of copy number variations in 2,026 healthy individuals
IGSR: The International Genome Sample Resource
��software * ttp://www.bioinf.jku.at/software/cnmops/cnmops.html cn.MOPS: mixture of Poissons for discovering copy number variations in next generation sequencing data��software {{DEFAULTSORT:Copy Number Variation Molecular biology Genomics Human evolution