|
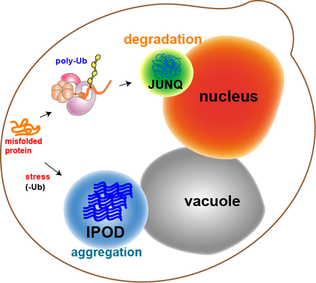
Protein Aggregation
In molecular biology, protein aggregation is a phenomenon in which intrinsically disordered proteins, intrinsically-disordered or mis-folded proteins aggregate (i.e., accumulate and clump together) either intra- or extracellularly. Protein aggregates have been implicated in a wide variety of diseases known as amyloidoses, including Amyotrophic lateral sclerosis, ALS, Alzheimer's, Parkinson's and prion disease. After synthesis, proteins typically Protein folding, fold into a particular Protein tertiary structure, three-dimensional conformation that is the most Thermodynamic equilibrium, thermodynamically favorable: their native state. This folding process is driven by the hydrophobic effect: a tendency for hydrophobic (water-fearing) portions of the protein to shield themselves from the hydrophilic (water-loving) environment of the cell by burying into the interior of the protein. Thus, the exterior of a protein is typically hydrophilic, whereas the interior is typically hydrophob ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alzheimer's
Alzheimer's disease (AD) is a neurodegenerative disease and the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As the disease advances, symptoms can include problems with language, disorientation (including easily getting lost), mood swings, loss of motivation, self-neglect, and behavioral issues. As a person's condition declines, they often withdraw from family and society. Gradually, bodily functions are lost, ultimately leading to death. Although the speed of progression can vary, the average life expectancy following diagnosis is three to twelve years. The causes of Alzheimer's disease remain poorly understood. There are many environmental and genetic risk factors associated with its development. The strongest genetic risk factor is from an allele of apolipoprotein E. Other risk factors include a history of head injury, clinical depression, and high blood pressure. The progression of the dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Aggregation
In molecular biology, protein aggregation is a phenomenon in which intrinsically disordered proteins, intrinsically-disordered or mis-folded proteins aggregate (i.e., accumulate and clump together) either intra- or extracellularly. Protein aggregates have been implicated in a wide variety of diseases known as amyloidoses, including Amyotrophic lateral sclerosis, ALS, Alzheimer's, Parkinson's and prion disease. After synthesis, proteins typically Protein folding, fold into a particular Protein tertiary structure, three-dimensional conformation that is the most Thermodynamic equilibrium, thermodynamically favorable: their native state. This folding process is driven by the hydrophobic effect: a tendency for hydrophobic (water-fearing) portions of the protein to shield themselves from the hydrophilic (water-loving) environment of the cell by burying into the interior of the protein. Thus, the exterior of a protein is typically hydrophilic, whereas the interior is typically hydrophob ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Disulfide
In chemistry, a disulfide (or disulphide in British English) is a compound containing a functional group or the anion. The linkage is also called an SS-bond or sometimes a disulfide bridge and usually derived from two thiol groups. In inorganic chemistry, the anion appears in a few rare minerals, but the functional group has tremendous importance in biochemistry. Disulfide bridges formed between thiol groups in two cysteine residues are an important component of the tertiary and quaternary structure of proteins. Compounds of the form are usually called ''persulfides'' instead. Organic disulfides Structure Disulfides have a C–S–S–C dihedral angle approaching 90°. The S–S bond length is 2.03 Å in diphenyl disulfide, similar to that in elemental sulfur. Disulfides are usually symmetric but they can also be unsymmetric. Symmetrical disulfides are compounds of the formula . Most disulfides encountered in organosulfur chemistry are symmetrical disulfides. Unsymme ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligomer
In chemistry and biochemistry, an oligomer () is a molecule that consists of a few repeating units which could be derived, actually or conceptually, from smaller molecules, monomers.Quote: ''Oligomer molecule: A molecule of intermediate relative molecular mass, the structure of which essentially comprises a small plurality of units derived, actually or conceptually, from molecules of lower relative molecular mass.'' The name is composed of Greek elements '' oligo-'', "a few" and '' -mer'', "parts". An adjective form is ''oligomeric''. The oligomer concept is contrasted to that of a polymer, which is usually understood to have a large number of units, possibly thousands or millions. However, there is no sharp distinction between these two concepts. One proposed criterion is whether the molecule's properties vary significantly with the removal of one or a few of the units. An oligomer with a specific number of units is referred to by the Greek prefix denoting that number, w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteolysis
Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Protein degradation is a major regulatory mechanism of gene expression and contributes substantially to shaping mammalian proteomes. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion. Proteolysis in organisms serves many purposes; for example, digestive enzymes break down proteins in food to provide amino acids for the organism, while proteolytic processing of a polypeptide chain after its synthesis may be necessary for the production of an active protein. It is also important in the regulation of some physiological and cellular processes including apoptosis, as well as preventing the accumulation of unwanted or misfolded proteins in cells. Consequently, abnormality in the regulation of proteolysis can cause diseases. Proteolysis can also ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
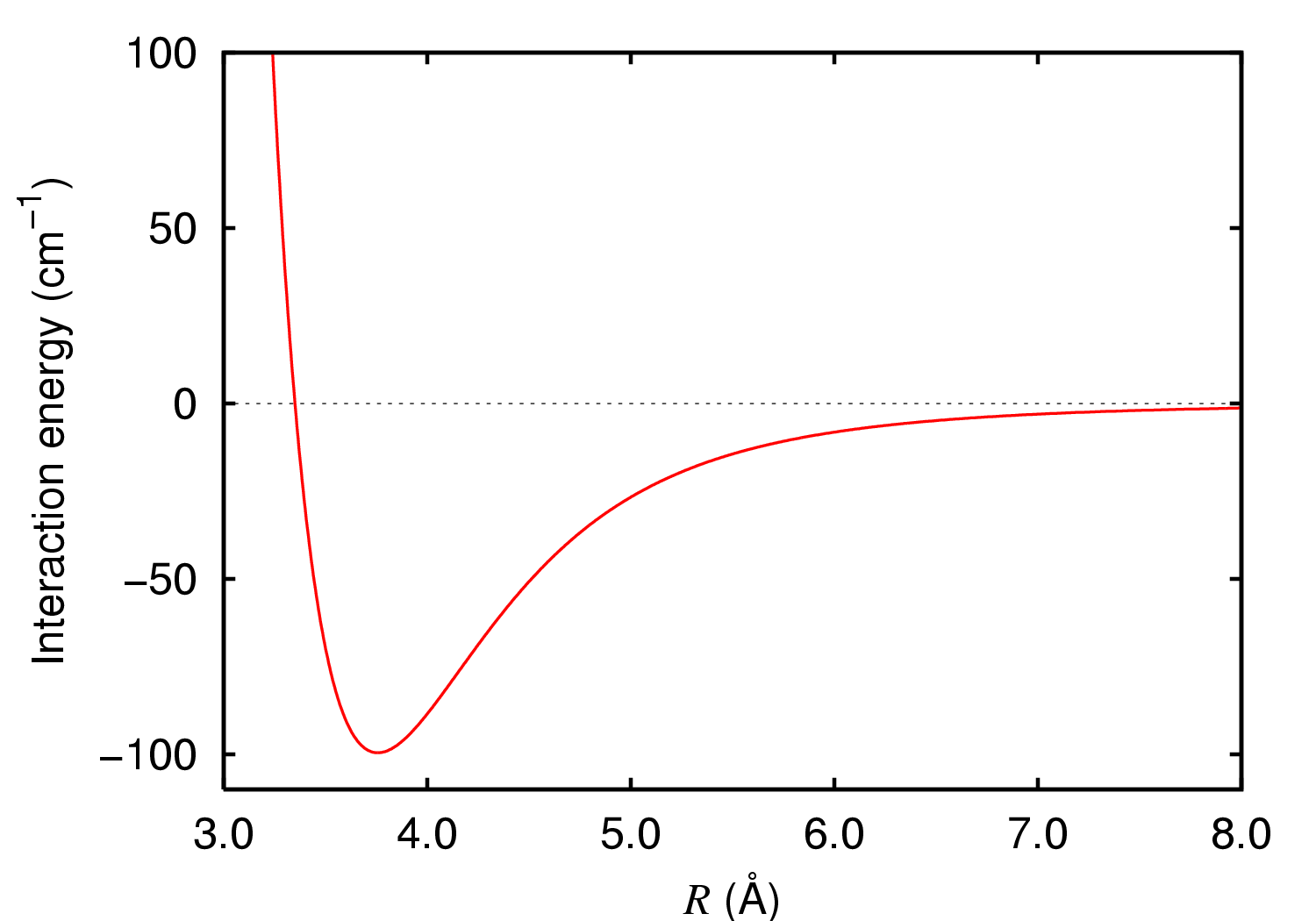
Dipole-dipole Bond
An intermolecular force (IMF; also secondary force) is the force that mediates interaction between molecules, including the electromagnetic forces of attraction or repulsion which act between atoms and other types of neighbouring particles (e.g. atoms or ions). Intermolecular forces are weak relative to intramolecular forces – the forces which hold a molecule together. For example, the covalent bond, involving sharing electron pairs between atoms, is much stronger than the forces present between neighboring molecules. Both sets of forces are essential parts of force fields frequently used in molecular mechanics. The first reference to the nature of microscopic forces is found in Alexis Clairaut's work ''Théorie de la figure de la Terre,'' published in Paris in 1743. Other scientists who have contributed to the investigation of microscopic forces include: Laplace, Gauss, Maxwell, Boltzmann and Pauling. Attractive intermolecular forces are categorized into the following types: * ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
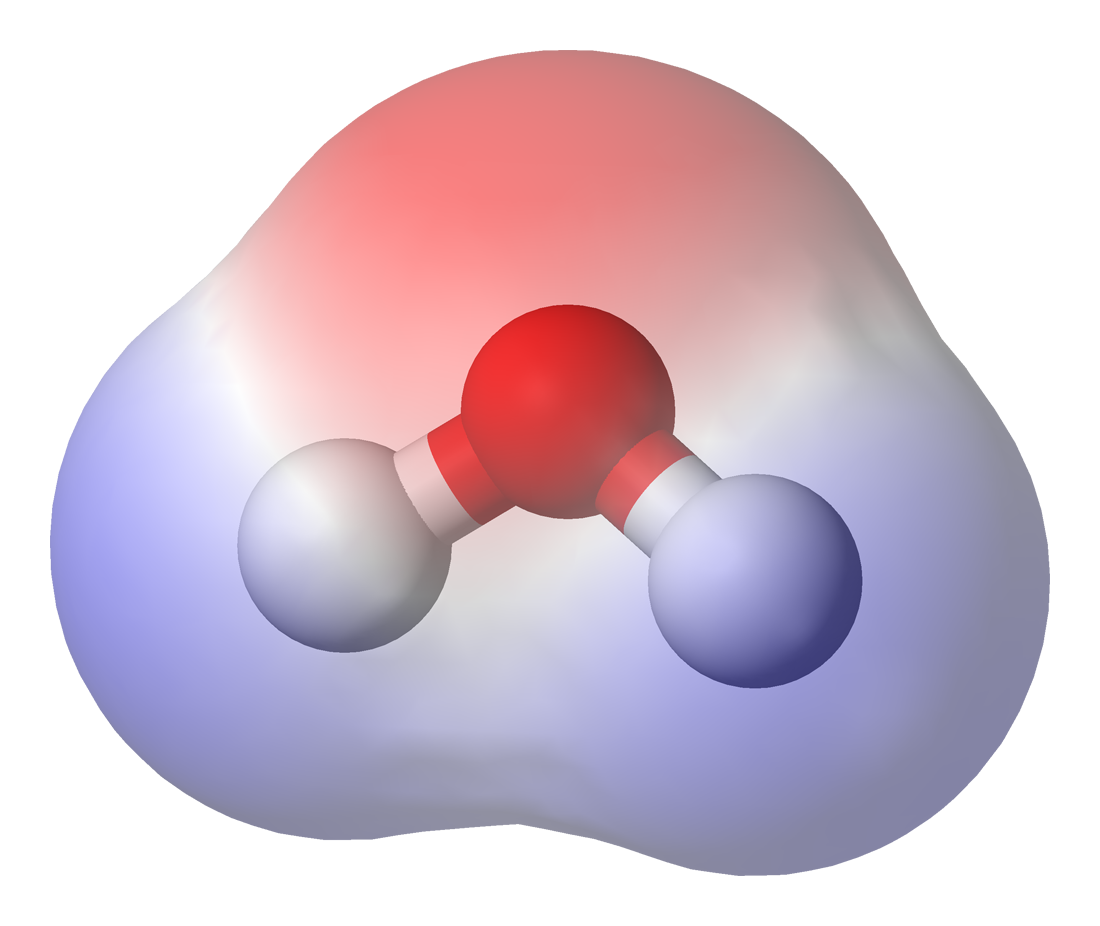
Hydrogen Bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently bonded to a more Electronegativity, electronegative donor atom or group (Dn), interacts with another electronegative atom bearing a lone pair of electrons—the hydrogen bond acceptor (Ac). Unlike simple Dipole–dipole attraction, dipole–dipole interactions, hydrogen bonding arises from charge transfer (nB → σ*AH), Atomic orbital, orbital interactions, and quantum mechanical Delocalized electron, delocalization, making it a resonance-assisted interaction rather than a mere electrostatic attraction. The general notation for hydrogen bonding is Dn−H···Ac, where the solid line represents a polar covalent bond, and the dotted or dashed line indicates the hydrogen bond. The most frequent donor and acceptor atoms are nitrogen (N), oxyg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
London Dispersion Force
London dispersion forces (LDF, also known as dispersion forces, London forces, instantaneous dipole–induced dipole forces, fluctuating induced dipole bonds or loosely as van der Waals forces) are a type of intermolecular force acting between atoms and molecules that are normally electrically symmetric; that is, the electrons are symmetrically distributed with respect to the nucleus. They are part of the van der Waals forces. The LDF is named after the German physicist Fritz London. They are the weakest of the intermolecular forces. Introduction The electron distribution around an atom or molecule undergoes fluctuations in time. These fluctuations create instantaneous electric fields which are felt by other nearby atoms and molecules, which in turn adjust the spatial distribution of their own electrons. The net effect is that the fluctuations in electron positions in one atom induce a corresponding redistribution of electrons in other atoms, such that the electron motions be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nonpolar
In chemistry, polarity is a separation of electric charge leading to a molecule or its chemical groups having an electric dipole moment, with a negatively charged end and a positively charged end. Polar molecules must contain one or more polar bonds due to a difference in electronegativity between the bonded atoms. Molecules containing polar bonds have no molecular polarity if the bond dipoles cancel each other out by symmetry. Polar molecules interact through dipole-dipole intermolecular forces and hydrogen bonds. Polarity underlies a number of physical properties including surface tension, solubility, and melting and boiling points. Polarity of bonds Not all atoms attract electrons with the same force. The amount of "pull" an atom exerts on its electrons is called its electronegativity. Atoms with high electronegativitiessuch as fluorine, oxygen, and nitrogenexert a greater pull on electrons than atoms with lower electronegativities such as alkali metals and alkaline ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |