|
Virusoid
Virusoids are circular single-stranded RNA(s) dependent on viruses for replication and encapsidation. The genome of virusoids consists of several hundred (200–400) nucleotides and does not code for any proteins. Virusoids are essentially viroids that have been encapsulated by a helper virus coat protein. They are thus similar to viroids in their means of replication (rolling circle replication) and in their lack of genes, but they differ in that viroids do not possess a protein coat. Both virusoids and a few viroids encode a hammerhead ribozyme. Virusoids, while being studied in virology, are subviral particles rather than viruses. Since they depend on helper viruses, they are classified as satellites. Virusoids are listed in virological taxonomy as Satellites/Satellite nucleic acids/Subgroup 3: Circular satellite RNA(s). Definition Depending on whether a lax or strict definition is used, the term ''virusoid'' may also include Hepatitis D virus (HDV). Like plant vir ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hammerhead Ribozyme
The hammerhead ribozyme is an RNA Sequence motif, motif that catalyzes reversible cleavage and Ligation (molecular biology), ligation reactions at a specific site within an RNA molecule. It is one of several catalytic RNAs (ribozymes) known to occur in nature. It serves as a model system for research on the Nucleic acid structure, structure and properties of RNA, and is used for targeted RNA cleavage experiments, some with proposed therapeutic applications. Named for the resemblance of early Nucleic acid secondary structure, secondary structure diagrams to a hammerhead shark, hammerhead ribozymes were originally discovered in two classes of plant virus-like RNAs: Satellite (biology), satellite RNAs and viroids. They are also known in some classes of retrotransposons, including the retrozymes. The hammerhead ribozyme motif has been ubiquitously reported in lineages across the Tree of life (biology), tree of life. The self-cleavage reactions, first reported in 1986, are part of a ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
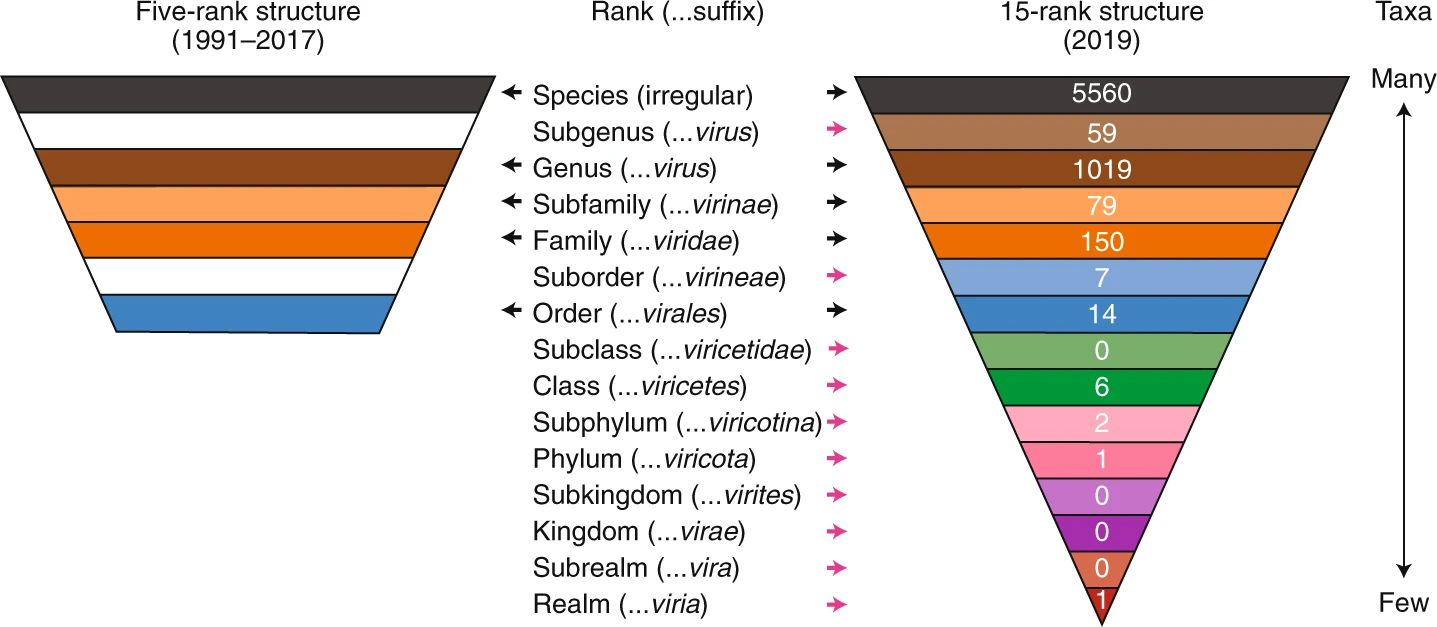
Virus Classification
Virus classification is the process of naming viruses and placing them into a taxonomy (biology), taxonomic system similar to the classification systems used for cell (biology), cellular organisms. Viruses are classified by phenotypic characteristics, such as Virus#Structure, morphology, nucleic acid type, mode of replication, Host (biology), host organisms, and the type of disease they cause. The formal taxonomic classification of viruses is the responsibility of the International Committee on Taxonomy of Viruses (ICTV) system, although the Baltimore classification system can be used to place viruses into one of seven groups based on their manner of mRNA synthesis. Specific naming conventions and further classification guidelines are set out by the ICTV. In 2021, the ICTV changed the International Code of Virus Classification and Nomenclature (ICVCN) to mandate a binomial format (genus, , , , species) for naming new viral species similar to that used for cellular organisms; the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Satellite (biology)
A satellite is a subviral agent that depends on the coinfection of a host cell with a helper virus for its replication. Satellites can be divided into two major groups: satellite viruses and satellite nucleic acids. Satellite viruses, which are most commonly associated with plants, are also found in mammals, arthropods, and bacteria. They encode structural proteins to enclose their genetic material, which are therefore distinct from the structural proteins of their helper viruses. Satellite nucleic acids, in contrast, do not encode their own structural proteins, but instead are encapsulated by proteins encoded by their helper viruses. The genomes of satellites range upward from 359 nucleotides in length for satellite tobacco ringspot virus RNA (STobRV). Most viruses have the capability to use host enzymes or their own replication machinery to independently replicate their own viral RNA. Satellites, in contrast, are completely dependent on a helper virus for replication. The sy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Viroid
Viroids are small single-stranded, circular RNAs that are infectious pathogens. Unlike viruses, they have no protein coating. All known viroids are inhabitants of angiosperms (flowering plants), and most cause diseases, whose respective economic importance to humans varies widely. A recent metatranscriptomics study suggests that the host diversity of viroids and viroid-like elements is broader than previously thought and that it would not be limited to plants, encompassing even the prokaryotes. The first discoveries of viroids in the 1970s triggered the historically third major extension of the biosphere—to include smaller lifelike entities—after the discoveries in 1675 by Antonie van Leeuwenhoek (of the "subvisible" microorganisms) and in 1892–1898 by Dmitri Iosifovich Ivanovsky and Martinus Beijerinck (of the "submicroscopic" viruses). The unique properties of viroids have been recognized by the International Committee on Taxonomy of Viruses, in creating a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virus Classification
Virus classification is the process of naming viruses and placing them into a taxonomy (biology), taxonomic system similar to the classification systems used for cell (biology), cellular organisms. Viruses are classified by phenotypic characteristics, such as Virus#Structure, morphology, nucleic acid type, mode of replication, Host (biology), host organisms, and the type of disease they cause. The formal taxonomic classification of viruses is the responsibility of the International Committee on Taxonomy of Viruses (ICTV) system, although the Baltimore classification system can be used to place viruses into one of seven groups based on their manner of mRNA synthesis. Specific naming conventions and further classification guidelines are set out by the ICTV. In 2021, the ICTV changed the International Code of Virus Classification and Nomenclature (ICVCN) to mandate a binomial format (genus, , , , species) for naming new viral species similar to that used for cellular organisms; the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Virus
A virus is a submicroscopic infectious agent that replicates only inside the living Cell (biology), cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 16,000 of the millions of List of virus species, virus species have been described in detail. The study of viruses is known as virology, a subspeciality of microbiology. When infected, a host cell is often forced to rapidly produce thousands of copies of the original virus. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent viral particles, or ''virions'', consisting of (i) genetic material, i.e., long ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hammerhead Ribozyme Ribbons
Hammerhead may refer to: * The head of a hammer Commercial * Hammerhead (company), a subsidiary of SRAM Corporation Fiction * Hammerhead (comics), a Marvel Comics foe of Spider-Man * ''Hammerhead'' (film), a 1968 film based on the novel by James Mayo * '' Hammerhead: Shark Frenzy'' a 2005 TV movie starring William Forsythe, Jeffery Combs and Hunter Tylo * ''Hammerhead'' (1987 film), a 1987 Italian action film directed by Enzo G. Castellari * ''Hammerhead'' (novel), a 1964 Charles Hood secret agent novel by James Mayo * Hammerhead Hannigan, the leader of Taurus Bulba's henchmen in the television cartoon series ''Darkwing Duck'' * Hammerhead, a Rulon character from the TV cartoon ''Dino-Riders'' * Hammerhead, the nickname for the ''Star Wars'' character Momaw Nadon * ''Hammerheads'', a 1990 book by Dale Brown * '' James Bond 007: Hammerhead'', a 2016 James Bond comic book by Dynamite Entertainment Games and rides * Hammerhead, a heavy helicopter gunship in the video g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SnRNP
snRNPs (pronounced "snurps"), or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs. The action of snRNPs is essential to the removal of introns from pre-mRNA, a critical aspect of post-transcriptional modification of RNA, occurring only in the nucleus of eukaryotic cells. Additionally, '' U7 snRNP'' is not involved in splicing at all, as U7 snRNP is responsible for processing the 3′ stem-loop of histone pre-mRNA. The two essential components of snRNPs are protein molecules and RNA. The RNA found within each snRNP particle is known as ''small nuclear RNA'', or snRNA, and is usually about 150 nucleotides in length. The snRNA component of the snRNP gives specificity to individual introns by " recognizing" the sequences of critical splicing signals at the 5' and 3' ends and branch site of introns. The snRNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Self-splicing Intron
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA, tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains – the P4-P6 domain (formed from the stacking of P5, P4, P6 and P6a helices) and the P3-P9 domain (formed from the P8, P3, P7 and P9 helices). The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions. Catalysis Splicing of group I introns is processed by two sequential transesterification reactions. First an exogenous guanosine or guanosine nucleotide (''exoG'') docks onto the active G-binding site located in P7, and then its 3'-OH is aligned to attack the phosphodiester bond at the "upstream" (closer to the 5' end) splice site located in P1, resulting in a free 3'-OH group at the upstream exon and the exoG bein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while many, typically called '' restriction endonucleases'' or ''restriction enzymes'', cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (''endo'') portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the ''restriction site''. Typically, a restriction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrahymena Thermophila
''Tetrahymena thermophila'' is a species of Ciliophora in the family Tetrahymenidae. It is a free living protozoon and occurs in fresh water. There is little information on the ecology and natural history of this species, but it is the most widely known and widely studied species in the genus '' Tetrahymena''. The species has been used as a model organism for molecular and cellular biology. It has also helped in the discovery of new genes as well as helping to understand the mechanisms of function of certain genes. Studies on this species have contributed to major discoveries in biology. For example, the MAT locus found in this species has provided a foundation for the evolution of mating systems. The species was at first considered to be a form of '' Tetrahymena pyriformis''. '' T. malaccensis'' is the closest relative to''T. thermophila''. Characteristics It is about 50 μm long. One famous trait this species is known for is that has 7 different mating types, unlike ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |