|
Staphylococcal Nuclease
Micrococcal nuclease (, ''S7 Nuclease'', ''MNase'', ''spleen endonuclease'', ''thermonuclease'', ''nuclease T'', ''micrococcal endonuclease'', ''nuclease T, ''staphylococcal nuclease'', ''spleen phosphodiesterase'', ''Staphylococcus aureus nuclease'', ''Staphylococcus aureus nuclease B'', ''ribonucleate (deoxynucleate) 3'-nucleotidohydrolase'') is an endo-exonuclease that preferentially digests single-stranded nucleic acids. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates. The enzyme is also active against double-stranded DNA and RNA and all sequences will be ultimately cleaved. Characteristics The enzyme has a molecular weight of 16.9kDa. The pH optimum is reported as 9.2. The enzyme activity is strictly dependent on Ca2+ and the pH optimum varies according to Ca2+ concentration. The enzyme is therefore easily inactivated by EGTA. Source ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning. There are two primary classifications based on the locus of activity. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the ''middle'' of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. History In the late 1960s, scientists Stuart Linn and Werner Arber isolated examples of the two types of enzymes responsible for phage growth restriction in Escherichi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reticulocyte
In hematology, reticulocytes are immature red blood cells (RBCs). In the process of erythropoiesis (red blood cell formation), reticulocytes develop and mature in the bone marrow and then circulate for about a day in the blood stream before developing into mature red blood cells. Like mature red blood cells, in mammals, reticulocytes do not have a cell nucleus. They are called reticulocytes because of a reticular (mesh-like) network of ribosomal RNA that becomes visible under a microscope with certain stains such as new methylene blue and Romanowsky stain. Clinical significance To accurately measure reticulocyte counts, automated counters use a combination of laser excitation, detectors and a fluorescent dye that marks RNA and DNA (such as titan yellow or polymethine). Reticulocytes appear slightly bluer than other red cells when looked at with the normal Romanowsky stain. Reticulocytes are also relatively large, a characteristic that is described by the mean corpuscular ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CUT&RUN Sequencing
CUT&RUN sequencing, also known as cleavage under targets and release using nuclease, is a method used to analyze protein interactions with DNA. CUT&RUN sequencing combines antibody-targeted controlled cleavage by micrococcal nuclease with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global DNA binding sites precisely for any protein of interest. Currently, ChIP-Seq is the most common technique utilized to study protein–DNA relations, however, it suffers from a number of practical and economical limitations that CUT&RUN sequencing does not. Uses CUT&RUN sequencing can be used to examine gene regulation or to analyze transcription factor and other chromatin-associated protein binding. Protein-DNA interactions regulate gene expression and are responsible for many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis. CUT&RUN is an alternative to th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Classification Of Proteins
The Structural Classification of Proteins (SCOP) database is a largely manual classification of protein structural domains based on similarities of their structures and amino acid sequences. A motivation for this classification is to determine the evolutionary relationship between proteins. Proteins with the same shapes but having little sequence or functional similarity are placed in different superfamilies, and are assumed to have only a very distant common ancestor. Proteins having the same shape and some similarity of sequence and/or function are placed in "families", and are assumed to have a closer common ancestor. Similar to CATH and Pfam databases, SCOP provides a classification of individual structural domains of proteins, rather than a classification of the entire proteins which may include a significant number of different domains. The SCOP database is freely accessible on the internet. SCOP was created in 1994 in the Centre for Protein Engineering and the Lab ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
OB-fold
In molecular biology, the OB-fold (oligonucleotide/oligosaccharide-binding fold) is a small protein structural motif observed in different proteins that bind oligonucleotides or oligosaccharides. It was originally identified in 1993 in four unrelated proteins: staphylococcal nuclease, anticodon binding domain of aspartyl-tRNA synthetase, and the B-subunits of heat-labile enterotoxin and verotoxin-1. Since then it has been found in multiple proteins many of which are involved in genome stability. This fold is often described as a Greek key motif. Structure The OB-fold consists of a five-stranded β-sheet coiled to form a closed β-barrel, capped by an α-helix located at one end and a binding cleft at the other. The α-helix packs against the bottom layer of residues, roughly perpendicular to the barrel axis. The β-sheet structure protrudes beyond this layer and packs around the sides of the helix. The binding specificities of each OB-fold depend on the different length, sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet (β-sheet, also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
An alpha helix (or α-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix). The alpha helix is the most common structural arrangement in the Protein secondary structure, secondary structure of proteins. It is also the most extreme type of local structure, and it is the local structure that is most easily predicted from a sequence of amino acids. The alpha helix has a right-handed helix conformation in which every backbone amino, N−H group hydrogen bonds to the backbone carbonyl, C=O group of the amino acid that is four residue (biochemistry), residues earlier in the protein sequence. Other names The alpha helix is also commonly called a: * Pauling–Corey–Branson α-helix (from the names of three scientists who described its structure) * 3.613-helix because there are 3.6 amino acids in one ring, with 13 atoms being involved in the ring formed by the hydrogen bond (starting with amidic hydrogen and ending with carbonyl oxygen) Discovery ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribbon Diagram
Ribbon diagrams, also known as Richardson diagrams, are three-dimensional space, 3D schematic representations of protein structure and are one of the most common methods of protein depiction used today. The ribbon depicts the general course and organization of the protein backbone in 3D and serves as a visual framework for hanging details of the entire atomic structure, such as the balls for the oxygen atoms attached to myoglobin's active site in the adjacent figure. Ribbon diagrams are generated by interpolating a smooth curve through the polypeptide backbone. Alpha helix, α-helices are shown as coiled ribbons or thick tubes, Beta sheet, β-sheets as arrows, and non-repetitive coils or loops as lines or thin tubes. The direction of the Peptide, polypeptide chain is shown locally by the arrows, and may be indicated overall by a colour ramp along the length of the ribbon. Ribbon diagrams are simple yet powerful, expressing the visual basics of a molecular structure (twist, fold an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, Nuclear magnetic resonance spectroscopy of proteins, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert Biocuration, biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organizations (PDBe, PDBj, RCSB PDB, and BMRB). The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
X-ray Crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to Diffraction, diffract in specific directions. By measuring the angles and intensities of the X-ray diffraction, a crystallography, crystallographer can produce a three-dimensional picture of the density of electrons within the crystal and the positions of the atoms, as well as their chemical bonds, crystallographic disorder, and other information. X-ray crystallography has been fundamental in the development of many scientific fields. In its first decades of use, this method determined the size of atoms, the lengths and types of chemical bonds, and the atomic-scale differences between various materials, especially minerals and alloys. The method has also revealed the structure and function of many biological molecules, including vitamins, drugs, proteins and nucleic acids such as DNA. X-ray crystall ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
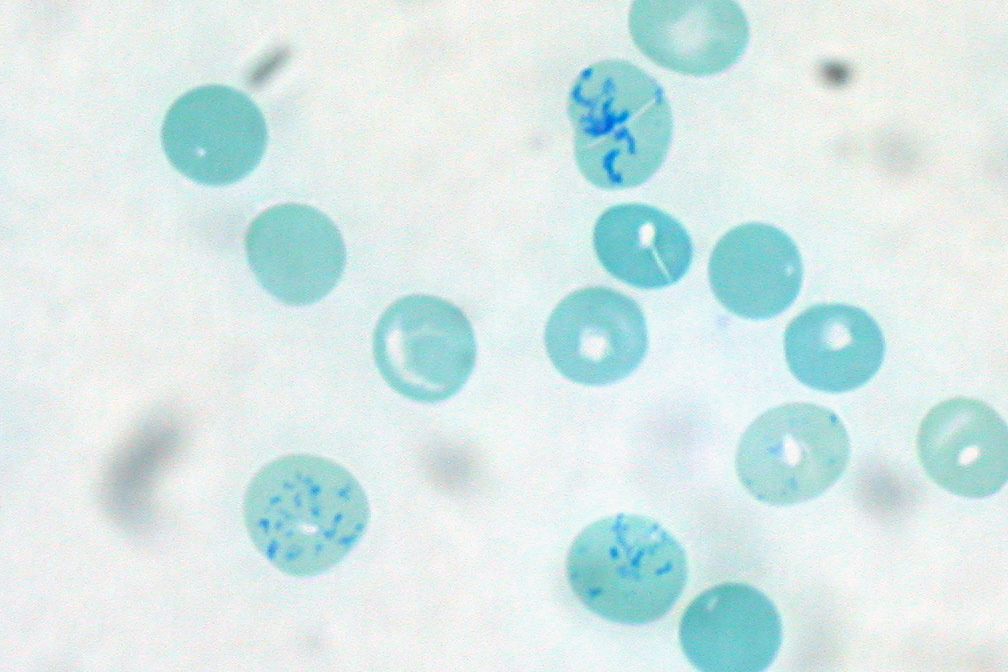
Staphylococcus
''Staphylococcus'', from Ancient Greek σταφυλή (''staphulḗ''), meaning "bunch of grapes", and (''kókkos''), meaning "kernel" or " Kermes", is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical ( cocci), and form in grape-like clusters. ''Staphylococcus'' species are facultative anaerobic organisms (capable of growth both aerobically and anaerobically). The name was coined in 1880 by Scottish surgeon and bacteriologist Alexander Ogston (1844–1929), following the pattern established five years earlier with the naming of '' Streptococcus''. It combines the prefix "staphylo-" (from ), and suffixed by the (from ). Staphylococcus was one of the leading infections in hospitals and many strains of this bacterium have become antibiotic resistant. Despite strong attempts to get rid of them, staphylococcus bacteria stay present in hospitals, where they can infect people who are most at ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |