|
Protein-L-isoaspartate(D-aspartate) O-methyltransferase
Protein L-isoaspartyl methyltransferase (PIMT, PCMT), also called S-adenosyl-L-methionine:protein-L-isoaspartate O-methyltransferase, is an enzyme which recognizes and catalyzes the repair of damaged L-isoaspartyl and D-aspartyl groups in proteins. It is a highly conserved enzyme which is present in nearly all eukaryotes, archaebacteria, and Gram-negative eubacteria. Function PIMT acts to transfer methyl groups from ''S''-adenosyl-L-methionine to the alpha side chain carboxyl groups of damaged L-isoaspartyl and D-aspartyl amino acids. The enzyme takes the end methyl residue from the methionine side chain and adds it to the side chain carboxyl group of L-isoaspartate or D-aspartate to create a methyl ester. Subsequent nonenzymatic reactions result in a rapid transformation to L-succinimide, which is a precursor to aspartate and isoaspartate. The L-succinimide can then undergo nonenzymatic hydrolysis, which generates some repaired L-aspartyl residues as well as some L-isoaspartyl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
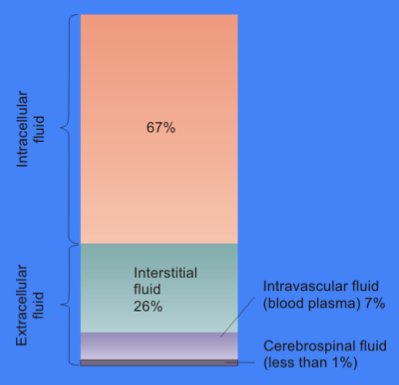
Cytosol
The cytosol, also known as cytoplasmic matrix or groundplasm, is one of the liquids found inside cells ( intracellular fluid (ICF)). It is separated into compartments by membranes. For example, the mitochondrial matrix separates the mitochondrion into many compartments. In the eukaryotic cell, the cytosol is surrounded by the cell membrane and is part of the cytoplasm, which also comprises the mitochondria, plastids, and other organelles (but not their internal fluids and structures); the cell nucleus is separate. The cytosol is thus a liquid matrix around the organelles. In prokaryotes, most of the chemical reactions of metabolism take place in the cytosol, while a few take place in membranes or in the periplasmic space. In eukaryotes, while many metabolic pathways still occur in the cytosol, others take place within organelles. The cytosol is a complex mixture of substances dissolved in water. Although water forms the large majority of the cytosol, its structure and prop ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methyltransferase
Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding ''S''-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Drosophila
''Drosophila'' () is a genus of flies, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or (less frequently) pomace flies, vinegar flies, or wine flies, a reference to the characteristic of many species to linger around overripe or rotting fruit. They should not be confused with the Tephritidae, a related family, which are also called fruit flies (sometimes referred to as "true fruit flies"); tephritids feed primarily on unripe or ripe fruit, with many species being regarded as destructive agricultural pests, especially the Mediterranean fruit fly. One species of ''Drosophila'' in particular, '' D. melanogaster'', has been heavily used in research in genetics and is a common model organism in developmental biology. The terms "fruit fly" and "''Drosophila''" are often used synonymously with ''D. melanogaster'' in modern biological literature. The entire genus, however, contains more than 1,500 species and is very diverse in appea ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate ( binding site) and residues that catalyse a reaction of that substrate (catalytic site). Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes. Each active site is evolved to be optimised to bind a particular substrate and catalyse a particular reaction, resulting in high specificity. This specificity is determined by the arrangement of amino acids within the active site and the structure of the substrates. Sometimes enzymes also need to bind with some cofactors to fulfil their function. The a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoleucine
Isoleucine (symbol Ile or I) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO form under biological conditions), and a hydrocarbon side chain with a branch (a central carbon atom bound to three other carbon atoms). It is classified as a non-polar, uncharged (at physiological pH), branched-chain, aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it, and must be ingested in our diet. Isoleucine is synthesized from pyruvate employing leucine biosynthesis enzymes in other organisms such as bacteria. It is encoded by the codons AUU, AUC, and AUA. Metabolism Biosynthesis As an essential nutrient, it is not synthesized in the body, hence it must be ingested, usually as a component of proteins. In plants and microorganisms, it is synthesized via several steps, sta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Valine
Valine (symbol Val or V) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated −NH3+ form under biological conditions), an α- carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain isopropyl group, making it a non-polar aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Human dietary sources are foods that contain protein, such as meats, dairy products, soy products, beans and legumes. It is encoded by all codons starting with GU (GUU, GUC, GUA, and GUG). History and etymology Valine was first isolated from casein in 1901 by Hermann Emil Fischer. The name valine comes from valeric acid, which in turn is named after the plant valerian due to the presence of the acid in the roots of the plant. Nomenclature According to IUPAC, carbon atoms forming valine are numbered sequenti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymorphism (biology)
In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative ''phenotypes'', in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population (one with random mating). Ford E.B. 1965. ''Genetic polymorphism''. Faber & Faber, London. Put simply, polymorphism is when there are two or more possibilities of a trait on a gene. For example, there is more than one possible trait in terms of a jaguar's skin colouring; they can be light morph or dark morph. Due to having more than one possible variation for this gene, it is termed 'polymorphism'. However, if the jaguar has only one possible trait for that gene, it would be termed "monomorphic". For example, if there was only one possible skin colour that a jaguar could have, it would be termed monomorphic. The term polyphenism can be used to clarify that the different forms arise from the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alternative Splicing
Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs will contain differences in their amino acid sequence and, often, in their biological functions (see Figure). Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome. In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene but many scientists believe that most o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GTPgammaS
GTPgammaS (GTPγS, guanosine 5'-O- amma-thioriphosphate) is a non-hydrolyzable or slowly hydrolyzable G-protein-activating analog of guanosine triphosphate (GTP). Many GTP binding proteins demonstrate activity when bound to GTP, and are inactivated via the hydrolysis of the phosphoanhydride bond that links the γ-phosphate to the remainder of the nucleotide, leaving a bound guanosine diphosphate (GDP) and releasing an inorganic phosphate. This usually occurs rapidly, and the GTP-binding protein can then only be activated by exchanging the GDP for a new GTP molecule. The substitution of sulfur for one of the oxygens of the γ-phosphate of GTP creates a nucleotide that either cannot be hydrolyzed or is only slowly hydrolyzed. This prevents the GTP-binding proteins from being inactivated, and allows the cellular processes that they carry out when active to be more easily studied. The consequences of the constitutive activation of GTP-binding proteins include stimulation of pho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoaspartate
Isoaspartic acid (isoaspartate, isoaspartyl, β-aspartate) is an aspartic acid residue isomeric to the typical α peptide linkage. It is a β-amino acid, with the side chain carboxyl moved to the backbone. Such a change is caused by a chemical reaction in which the nitrogen atom on the N+1 following peptide bond (in black at top right of Figure 1) nucleophilically attacks the γ-carbon of the side chain of an asparagine or aspartic acid residue, forming a succinimide intermediate (in red). Hydrolysis of the intermediate results in two products, either aspartic acid (in black at left) or isoaspartic acid, which is a β-amino acid (in green at bottom right). The reaction also results in the deamidation of the asparagine residue. Racemization may occur leading to the formation of D-aminoacids. Kinetics of isoaspartyl formation Isoaspartyl formation reactions have been conjectured to be one of the factors that limit the useful lifetime of protein Proteins are large biomolecule ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |