|
Phosphatidylinositol-4-phosphate 3-kinase
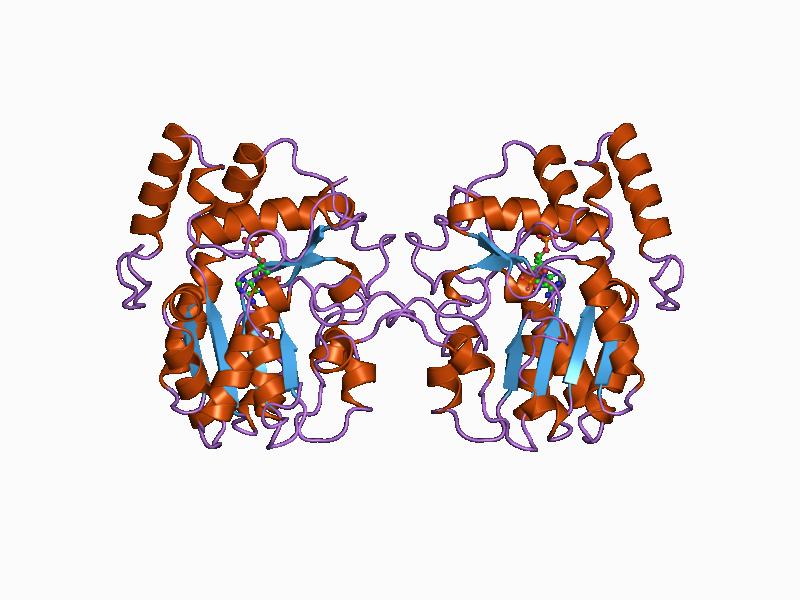
In enzymology, a phosphatidylinositol-4-phosphate 3-kinase () is an enzyme that catalyzes the chemical reaction :ATP + 1-phosphatidyl-1D-myo-inositol 4-phosphate \rightleftharpoons ADP + 1-phosphatidyl-1D-myo-inositol 3,4-bisphosphate Thus, the two substrates of this enzyme are ATP and 1-phosphatidyl-1D-myo-inositol 4-phosphate, whereas its two products are ADP and 1-phosphatidyl-1D-myo-inositol 3,4-bisphosphate. This enzyme belongs to the family of transferases, specifically those transferring phosphorus-containing groups (phosphotransferases) with an alcohol group as acceptor. The systematic name of this enzyme class is ATP:1-phosphatidyl-1D-myo-inositol-4-phosphate 3-phosphotransferase. Other names in common use include type II phosphoinositide 3-kinase, C2-domain-containing phosphoinositide 3-kinase, and phosphoinositide 3-kinase. This enzyme participates in phosphatidylinositol signaling system. Structural studies As of late 2007, 3 structures A structure is an arr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphatidylinositol 3,4-bisphosphate
Phosphatidylinositol (3,4)-bisphosphate (PtdIns(3,4)''P''2) is a minor phospholipid component of cell membranes, yet an important second messenger. The generation of PtdIns(3,4)''P''2 at the plasma membrane activates a number of important cell signaling pathways. Of all the phospholipids found within the membrane, inositol phospholipids make up less than 10%. Phosphoinositides (PIs), also known as phosphatidylinositol phosphates, are synthesized in the cell's endoplasmic reticulum by the protein phosphatidylinositol synthase (PIS). PIs are highly compartmentalized; their main components include a glycerol backbone, two fatty acid chains enriched with stearic acid and arachidonic acid, and an inositol ring whose phosphate groups' regulation differs between organelles depending on the specific PI and PIP kinases and PIP phosphatases present in the organelle. These kinases and phosphatases conduct phosphorylation and dephosphorylation at the inositol sugar head groups 3’, 4’, an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). This structural data is obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, Nuclear magnetic resonance spectroscopy of proteins, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert Biocuration, biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organizations (PDBe, PDBj, RCSB PDB, and BMRB). The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the backbone may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure. A number of these structures may bind to each other, forming a quaternary structure. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypeptide chains and amino acid side chains, it was Dorothy Maud Wrinch who incorporated geometry into the prediction of protein structures. Wrinch demon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphatidylinositol Signaling System
Phosphatidylinositol or inositol phospholipid is a biomolecule. It was initially called "inosite" when it was discovered by Léon Maquenne and Johann Joseph von Scherer in the late 19th century. It was discovered in bacteria but later also found in eukaryotes, and was found to be a signaling molecule. The biomolecule can exist in 9 different isomers. It is a lipid which contains a phosphate group, two fatty acid chains, and one inositol sugar molecule. Typically, the phosphate group has a negative charge (at physiological pH values). As a result, the molecule is amphiphilic. The production of the molecule is limited to the endoplasmic reticulum. History of phospatidylinositol Phosphatidylinositol (PI) and its derivatives have a rich history dating back to their discovery by Johann Joseph von Scherer and Léon Maquenne in the late 19th century. Initially known as "inosite" based on its sweet taste, the isolation and characterization of inositol laid the groundwork for unders ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
Enzymes are listed here by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system: :Oxidoreductases (EC 1) ( Oxidoreductase) * Dehydrogenase * Luciferase * DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) ** Homoserine dehydrogenase ** Aminopropanol oxidoreductase ** Diacetyl reductase ** Glycerol dehydrogenase ** Propanediol-phosphate dehydrogenase ** glycerol-3-phoshitiendopene dehydrogenase (NAD+) ** D-xylulose reductase ** L-xylulose reductase ** Lactate dehydrogenase ** Malate dehydrogenase ** Isocitrate dehydrogenase ** HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) ** Glucose oxidase ** L-gulonolactone oxidase ** Thiamine oxidase ** Xanthine oxidase * EC 1.1.4 (with a disulfide as accep ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phosphotransferase
In molecular biology, phosphotransferases are proteins in the transferase family of enzymes ( EC number 2.7) that catalyze certain chemical reactions. The general form of the phosphorylation reactions they catalyze is: \ce Where P is a phosphate group and A and B are the donating and accepting molecules, respectively. Classification Phosphotransferases are generally classified according to the acceptor molecule. , Classification in this article follows the rules of Enzyme Nomenclature of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). *EC 2.7.1 Phosphotransferases with an alcohol group as acceptor *EC 2.7.2 Phosphotransferases with a carb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transferase
In biochemistry, a transferase is any one of a class of enzymes that catalyse the transfer of specific functional groups (e.g. a methyl or glycosyl group) from one molecule (called the donor) to another (called the acceptor). They are involved in hundreds of different biochemical pathways throughout biology, and are integral to some of life's most important processes. Transferases are involved in myriad reactions in the cell. Three examples of these reactions are the activity of coenzyme A (CoA) transferase, which transfers thiol esters, the action of N-acetyltransferase, which is part of the pathway that metabolizes tryptophan, and the regulation of pyruvate dehydrogenase (PDH), which converts pyruvate to acetyl CoA. Transferases are also utilized during translation. In this case, an amino acid chain is the functional group transferred by a peptidyl transferase. The transfer involves the removal of the growing amino acid chain from the tRNA molecule in the A-site of the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Diphosphate
Adenosine diphosphate (ADP), also known as adenosine pyrophosphate (APP), is an important organic compound in metabolism and is essential to the flow of energy in living cells. ADP consists of three important structural components: a sugar backbone attached to adenine and two phosphate groups bonded to the 5 carbon atom of ribose. The diphosphate group of ADP is attached to the 5’ carbon of the sugar backbone, while the adenine attaches to the 1’ carbon. ADP can be interconverted to adenosine triphosphate (ATP) and adenosine monophosphate (AMP). ATP contains one more phosphate group than ADP, while AMP contains one fewer phosphate group. Energy transfer used by all living things is a result of dephosphorylation of ATP by enzymes known as ATPases. The cleavage of a phosphate group from ATP results in the coupling of energy to metabolic reactions and a by-product of ADP. ATP is continually reformed from lower-energy species ADP and AMP. The biosynthesis of ATP is achieved th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts include Ribozyme, catalytic RNA molecules, also called ribozymes. They are sometimes descr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Product (chemistry)
Products are the species formed from chemical reactions. During a chemical reaction, reactants are transformed into products after passing through a high energy transition state. This process results in the consumption of the reactants. It can be a spontaneous reaction or mediated by catalysts which lower the energy of the transition state, and by solvents which provide the chemical environment necessary for the reaction to take place. When represented in chemical equations, products are by convention drawn on the right-hand side, even in the case of reversible reactions. The properties of products such as their energies help determine several characteristics of a chemical reaction, such as whether the reaction is exergonic or endergonic. Additionally, the properties of a product can make it easier to extract and purify following a chemical reaction, especially if the product has a different state of matter than the reactants. Spontaneous reaction : R \rightarrow P *W ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
1-phosphatidyl-1D-myo-inositol 4-phosphate
Phosphatidylinositol-4-phosphate (PtdIns4''P'', PI-4-P, PI4P, or PIP) is a precursor of phosphatidylinositol (4,5)-bisphosphate. PtdIns4''P'' is prevalent in the membrane of the Golgi apparatus. In the Golgi apparatus, PtdIns4''P'' binds to the GTP-binding protein ARF and to effector proteins, including four-phosphate-adaptor protein 1 and 2 ( PLEKHA3 and PLEKHA8). This three molecule complex recruits proteins that need to be carried to the cell membrane. There is now evidence that PI-4-P is capable of deforming lipid systems into tightly curved assemblies, this is consistent with similar behaviour observed in phosphatidylinositol. See also * Phosphatidylinositol 3-phosphate * Phosphatidylinositol 5-phosphate * Phosphatidylinositol (3,4,5)-trisphosphate Phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)''P''3), abbreviated PIP3, is the product of the class I phosphoinositide 3-kinases' (PI 3-kinases) phosphorylation of phosphatidylinositol (4,5)-bisphosphate (PIP2). ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |